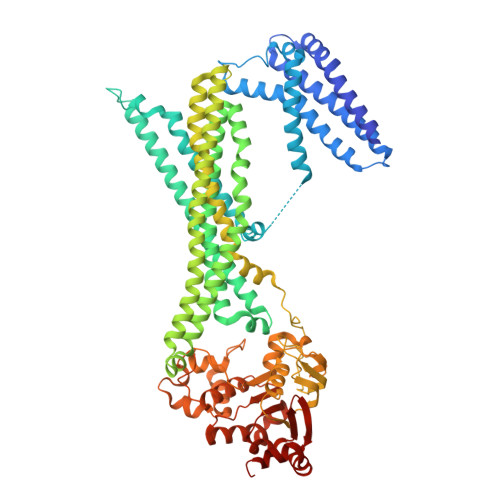
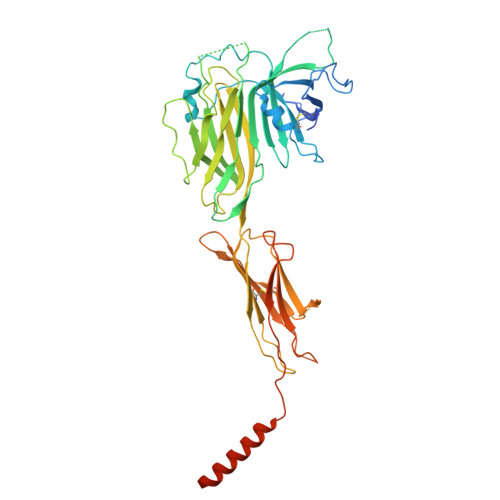
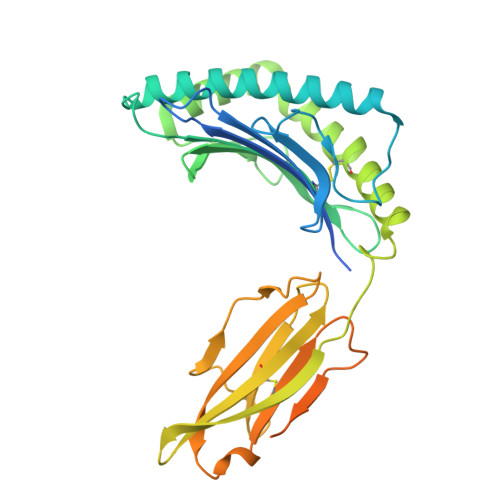
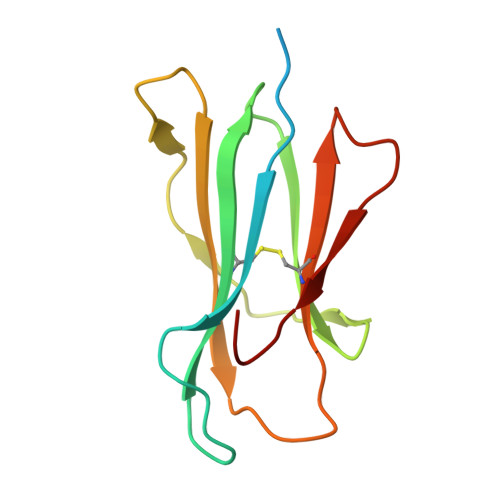
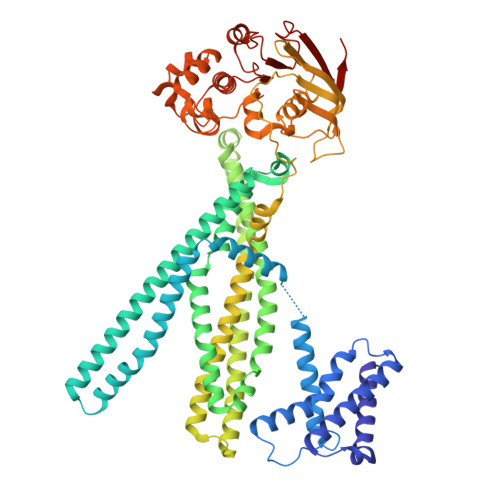
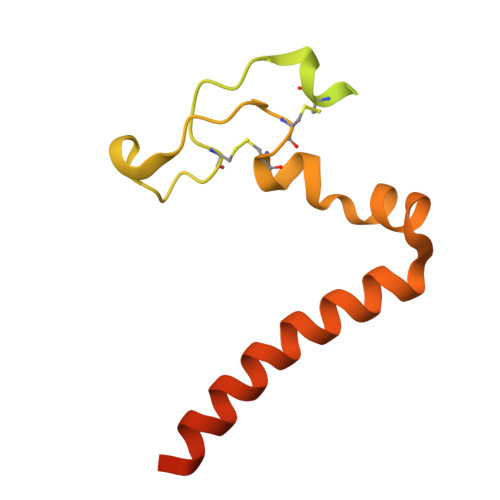
Architectural principles of transporter-chaperone coupling within the native MHC I peptide-loading complex.
Stolz, M., Susac, L., Fahim, A., Keller, R., Saggau, L., Mancia, F., Trowitzsch, S., Tampe, R.(2026) Sci Adv 12: eaea7735-eaea7735
- PubMed: 41481733
- DOI: https://doi.org/10.1126/sciadv.aea7735
- Primary Citation of Related Structures:
9RCV - PubMed Abstract:
Adaptive immunity depends on major histocompatibility complex class I (MHC I) presentation of peptides, a process orchestrated by the peptide-loading complex (PLC) in the endoplasmic reticulum (ER). The PLC ensures precise peptide selection and loading and is a major target of viral immune evasion, notably by human cytomegalovirus (HCMV). Here, we report the 2.59- to 2.88-Å cryo-electron microscopy structure of native human PLC bound to the HCMV immune evasin US6. US6 inhibits the transporter associated with antigen processing 1/2 (TAP1/2) by laterally attaching its transmembrane helix to TAP2 using a disulfide-rich domain to mimic a translocating peptide. This domain blocks the ER-lumenal exit and locks TAP in an outward-facing conformation with closed nucleotide-binding domains and asymmetric adenosine 5'-triphosphate/adenosine 5'-diphosphate occlusion. The structure also reveals how TAP's amino-terminal transmembrane domains scaffold the MHC I chaperone tapasin. These findings elucidate the mechanism of US6-mediated immune evasion and highlight potential targets for therapeutic modulation of immune presentation in infection and cancer.
- Institute of Biochemistry, Biocenter, Goethe University Frankfurt, Max-von-Laue-Str. 9, 60438 Frankfurt am Main, Germany.
Organizational Affiliation: