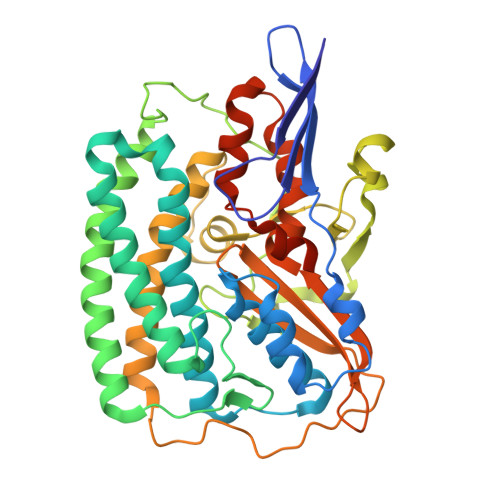
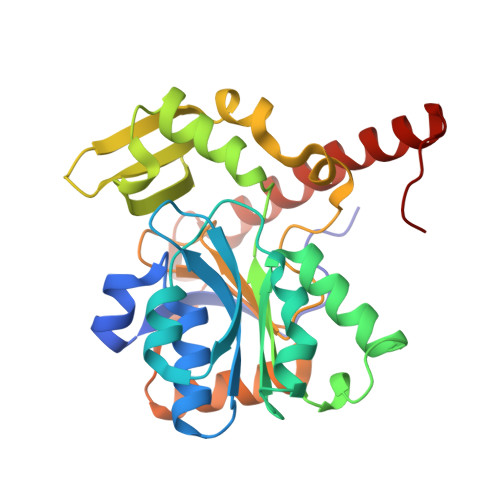
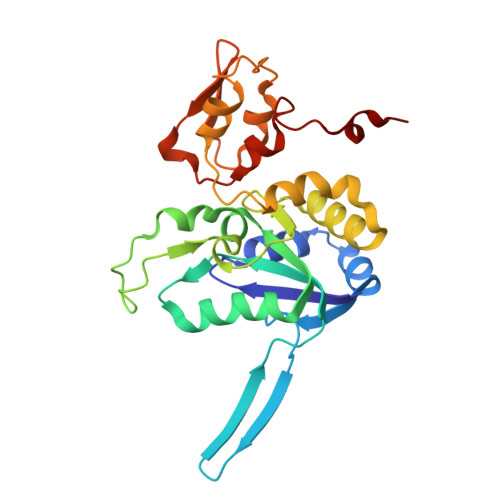
Structural and Spectroscopic Insights into Catalytic Intermediates of a [NiFe]-hydrogenase from Group 3.
Jespersen, M., Lorent, C., Lemaire, O.N., Zebger, I., Wagner, T.(2025) Chembiochem 26: e202500692-e202500692
- PubMed: 41078086
- DOI: https://doi.org/10.1002/cbic.202500692
- Primary Citation of Related Structures:
9R51, 9R52, 9R5I, 9R6Z - PubMed Abstract:
Hydrogenases catalyze reversible H 2 production and are potential models for renewable energy catalysts. Here, the full redox landscape of a group 3 [NiFe]-hydrogenase from methanothermococcus thermolithotrophicus is elucidated, resembling group 1 enzymes. Structural and spectroscopic analyses reveal a catalytic-ready state with nickel seesaw coordination, enabling intermediate trapping and advancing mechanistic understanding of oxygen-sensitive [NiFe] enzymes.
- Max Planck Institute for Marine Microbiology, Celsiusstraße 1, 28359, Bremen, Germany.
Organizational Affiliation: