Leveraging protein representations to explore uncharted fold spaces with generative models
Miao, Y., Pacesa, M., Georgeon, S., Schmidt, J., Lu, T., Huang, P.S., Correia, B.(2025) bioRxiv
Experimental Data Snapshot
Starting Model: in silico
View more details
wwPDB Validation 3D Report Full Report
(2025) bioRxiv
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| N37 | 147 | synthetic construct | Mutation(s): 0 | 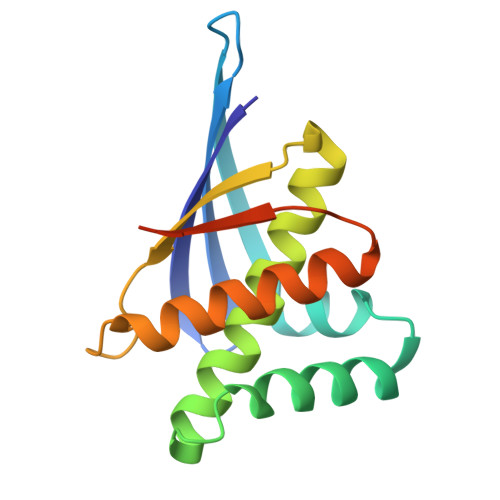 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ETA Query on ETA | I [auth A] | ETHANOLAMINE C2 H7 N O HZAXFHJVJLSVMW-UHFFFAOYSA-N |  | ||
| ACT Query on ACT | J [auth A] L [auth B] N [auth B] O [auth B] R [auth D] | ACETATE ION C2 H3 O2 QTBSBXVTEAMEQO-UHFFFAOYSA-M |  | ||
| SCN Query on SCN | AA [auth H] K [auth A] M [auth B] Q [auth C] S [auth D] | THIOCYANATE ION C N S ZMZDMBWJUHKJPS-UHFFFAOYSA-M |  | ||
| K Query on K | BA [auth H], P [auth B], V [auth E], X [auth F], Z [auth G] | POTASSIUM ION K NPYPAHLBTDXSSS-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 62.76 | α = 78.814 |
| b = 66.84 | β = 74.747 |
| c = 81.12 | γ = 62.558 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| autoPROC | data reduction |
| XSCALE | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Swiss National Science Foundation | Switzerland | -- |