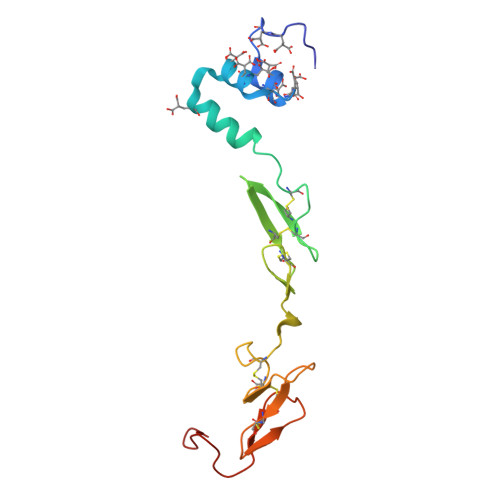
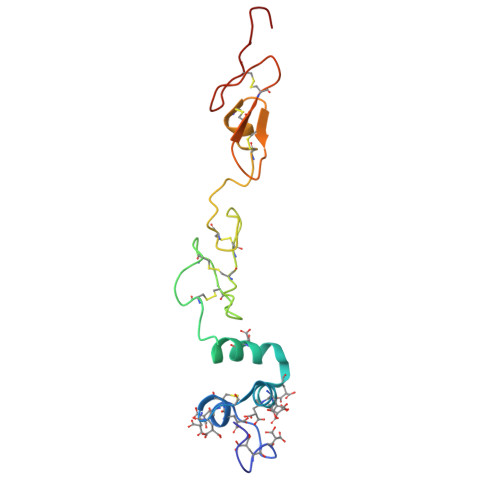
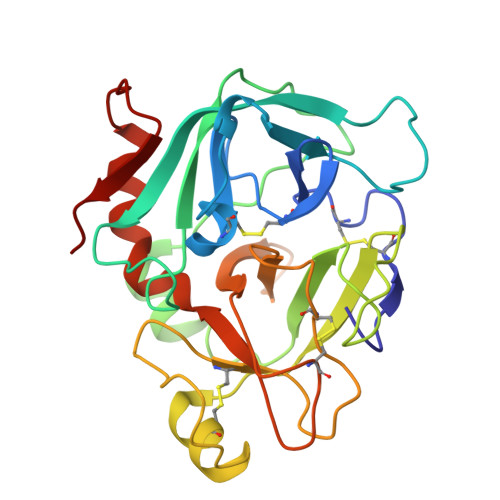
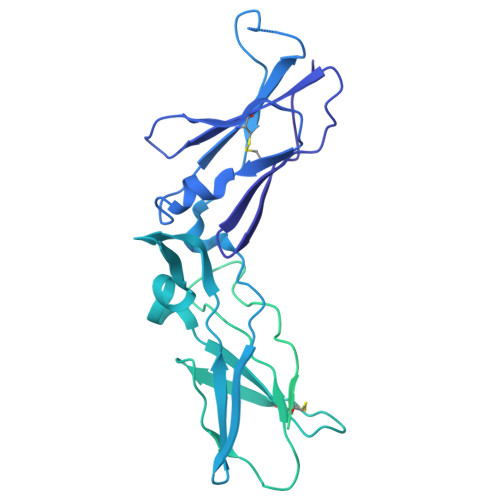
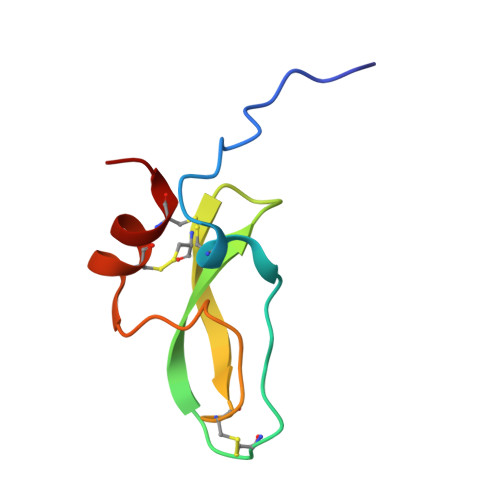
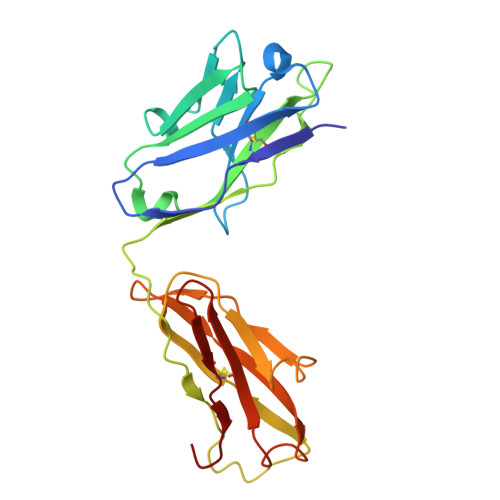
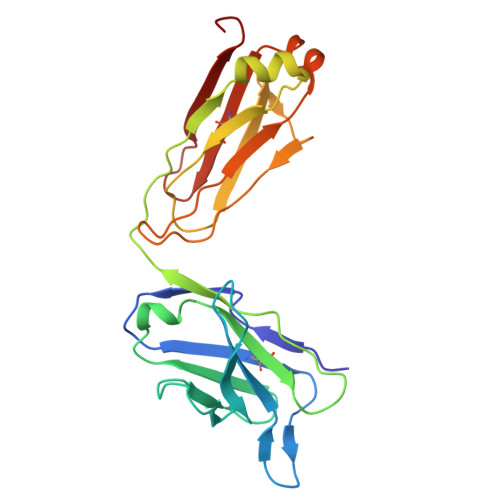
Cryo-EM structure of the tissue factor/factor VIIa complex with a factor X mimetic reveals a novel allosteric mechanism.
Sedzro, J.C., Photenhauer, A.L., Birkle, F., Meze, K., Mortenson, A., Duckworth, C., Wen, P.C., Kearns, S., Cianfrocco, M.A., Tajkhorshid, E., Ohi, M.D., Morrissey, J.H.(2025) Blood 146: 2833-2842
- PubMed: 40811856
- DOI: https://doi.org/10.1182/blood.2025029430
- Primary Citation of Related Structures:
9P0X - PubMed Abstract:
Blood clotting is triggered in hemostasis and thrombosis when the membrane-bound tissue factor (TF)/factor VIIa (FVIIa) complex activates factor X (FX). There are no structures of TF/FVIIa on membranes, with or without FX. Using cryo-EM to address this gap, we assembled TF/FVIIa complexes on nanoscale membrane bilayers (nanodiscs), bound to XK1 and an antibody fragment. XK1 is a FX mimetic whose protease domain is replaced by the first Kunitz-type (K1) domain of tissue factor pathway inhibitor, while 10H10 is a non-inhibitory, anti-TF antibody. We determined a cryo-EM structure of a TF/FVIIa/XK1/10H10/nanodisc complex with a resolution of 3.7 Å, allowing us to model all the protein backbones. TF/FVIIa extends perpendicularly from the membrane, interacting with a "handle shaped" XK1 at two locations: the K1 domain docks into FVIIa's active site, while the γ-carboxyglutamate-rich (GLA) domain binds to TF's substrate-binding exosite. The FX and FVIIa GLA domains also contact each other and the membrane surface. Except for a minor contact between the first epidermal growth factor (EGF)-like domain of XK1 and TF, the rest of the FX light chain does not interact with TF/FVIIa. The structure reveals a previously unrecognized, membrane-dependent allosteric activation mechanism between FVIIa and TF where a serine-rich loop in TF that partially obscures the TF exosite must undergo a shift to allow access of the FX GLA domain to its final binding location on the membrane-bound TF/FVIIa complex. This mechanism also provides a novel explanation for the otherwise puzzling phenomenon of TF encryption/decryption on cell surfaces.
- University of Michigan, Ann Arbor, Michigan, United States.
Organizational Affiliation: