NitrOFF: An Engineered Fluorescent Biosensor to Illuminate Nitrate Transport in Living Cells.
Cook, M.A., Smailys, J.D., Ji, K., Phelps, S.M., Tutol, J.N., Kim, W., Ong, W.S.Y., Peng, W., Maydew, C., Zhang, Y.J., Dodani, S.C.(2025) Angew Chem Int Ed Engl 64: e202508058-e202508058
- PubMed: 40856431
- DOI: https://doi.org/10.1002/anie.202508058
- Primary Citation of Related Structures:
9NCU, 9NCX - PubMed Abstract:
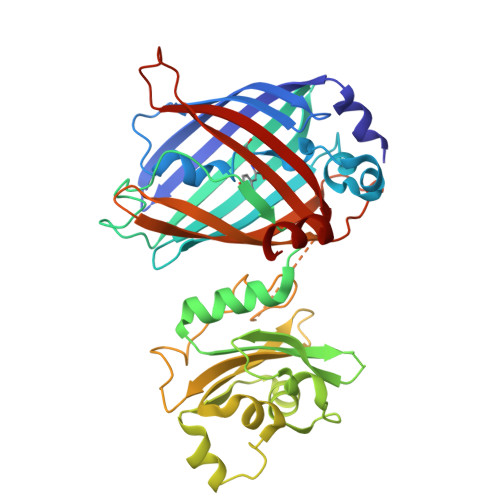
The duality of nitrate is nowhere better exemplified than in human physiology-a detrimental pollutant but also a protective nutrient-particularly as connected to nitric oxide. Aside from limited insights into nitrate uptake and storage, foundational nitrate biology has lagged. Genetically encoded fluorescent biosensors can address this gap with real-time imaging, but such technologies for mammalian cell applications remain rare. Here, we designed and engineered a biosensor fusing the green fluorescent protein EGFP and the nitrate recognition domain NreA from Staphylococcus carnosus. Seven rounds of directed evolution and 15 mutations resulted in NitrOFF. NitrOFF has a high degree of allosteric communication between the domains reflected in a turn-off intensiometric response (K d ≈ 9 µM). This was further reinforced by X-ray crystal structures of apo and nitrate-bound NitrOFF, which revealed a large-scale conformational rearrangement changing the relative positioning of the domains by 68.4°. This dramatic difference was triggered by the formation of a long helix at the engineered linker connecting the two domains, peeling the β7 strand off the EGFP and thus extinguishing the fluorescence upon nitrate binding. Finally, we highlighted the utility of NitrOFF to monitor exogenous nitrate uptake and modulation in the human embryonic kidney (HEK) 293 cell line.
- Department of Chemistry and Biochemistry, The University of Texas at Dallas, 800 West Campbell Road, Richardson, TX, 75080, USA.
Organizational Affiliation: