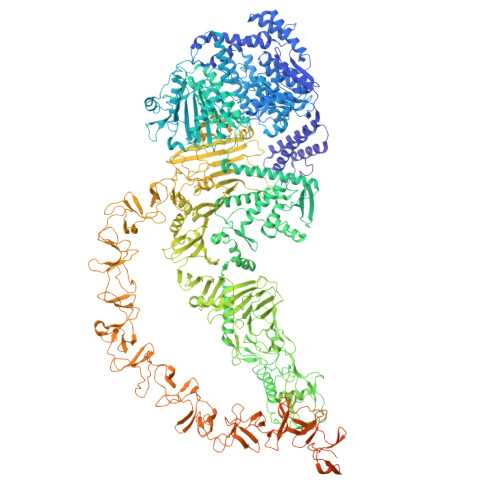
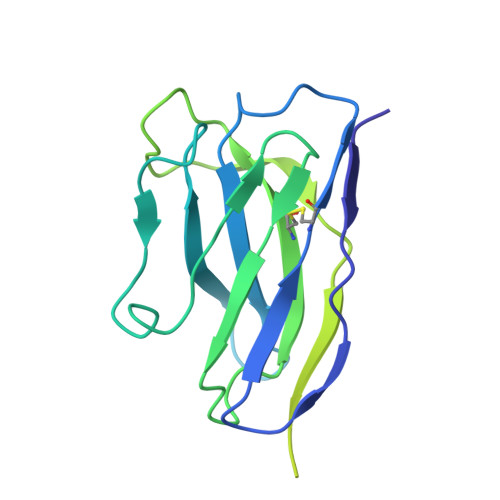
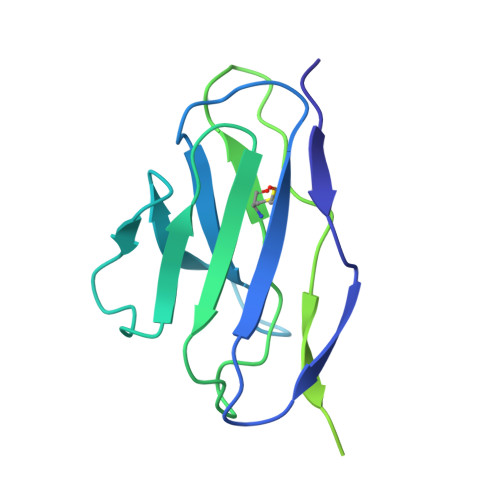
Mouse monoclonal antibodies against Clostridioides difficile toxins TcdA and TcdB target diverse epitopes for neutralization.
Kroh, H.K., Jensen, J.L., Wellnitz, S., Park, J.J., Esadze, A., Huynh, K.W., Ammirati, M., Han, S., Anderson, A.S., Lacy, D.B., Gribenko, A.(2025) Infect Immun 93: e0013925-e0013925
- PubMed: 40844461
- DOI: https://doi.org/10.1128/iai.00139-25
- Primary Citation of Related Structures:
9MX1 - PubMed Abstract:
Clostridioides difficile is a spore-forming, Gram-positive bacterium that can cause infections in subjects with weakened immune system or following antibiotic treatment. These infections may lead to pseudomembranous colitis and antibiotic-associated diarrhea in humans. As such, C. difficile is a major cause of nosocomial illness worldwide. Major virulence factors of the bacterium are the large clostridium toxins A (TcdA) and B (TcdB)-high molecular mass proteins with intrinsic glucosyltransferase activity. Toxins bind to the intestinal epithelium and undergo endocytosis by the epithelial cells, followed by a conformational change triggered by the low pH of early endosomes. This conformational change leads to the exposure of hydrophobic segments, followed by membrane insertion, formation of pores, and translocation of the glucosyltransferase domain into the cellular cytoplasm. Once in the cytoplasm, the glucosyltransferase domain inactivates small GTPases of the Rho family of proteins, leading to the disruption of the cytoskeleton. In the current work, we describe the discovery and characterization of a panel of neutralizing mouse monoclonal antibodies capable of interfering with several steps of cellular intoxication by the toxins. The antibodies were produced using hybridoma technology. Neutralizing activity of the antibodies was confirmed using toxin neutralization assays, and functional assays were used to identify specific neutralization mechanisms. Binding epitopes of the antibodies were identified by hydrogen-deuterium exchange mass spectrometry and confirmed through negative-stain and cryo-electron microscopy. Together, our results show that full-length toxins and/or genetically- and chemically-modified toxoids can induce a wide spectrum of antibodies capable of neutralizing the toxins via a variety of mechanisms.
- Department of Pathology, Microbiology, and Immunology, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
Organizational Affiliation: