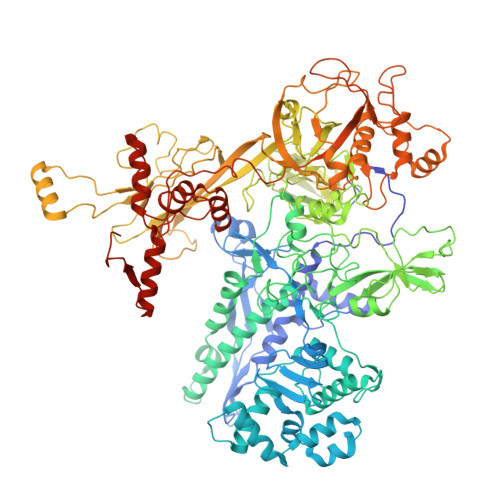
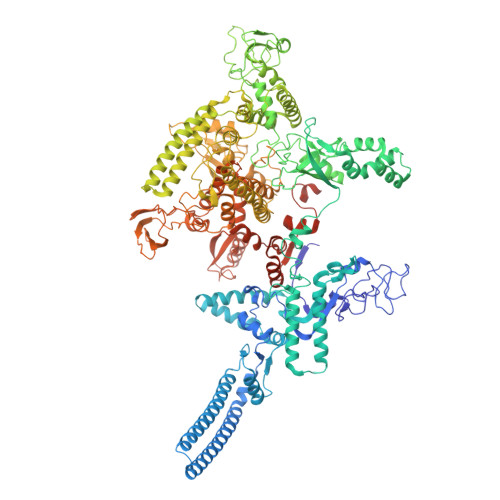
Molecular basis of Streptomyces ECF sigma ShbA factors transcribing principal sigma HrdB genes.
Liu, G., Yang, X., Yan, W., Wang, Y., Yu, F., Zheng, J.(2025) Nucleic Acids Res 53
- PubMed: 40272361
- DOI: https://doi.org/10.1093/nar/gkaf339
- Primary Citation of Related Structures:
9ISN, 9M84 - PubMed Abstract:
In bacteria, principal σ factors (σ70 or σA) transcribe housekeeping genes required for cell viability. Although most principal σ genes are transcribed by the RNA polymerase (RNAP) holoenzyme containing the principal σ factor itself, an extracytoplasmic function (ECF) σ factor (σShbA) governs transcription of the principal σ factor gene (hrdB) in two model Streptomycetes. Here, we employed a combination of cryo-electron microscopy (cryo-EM) and bioinformatics to decipher how σShbA-RNAP holoenzymes govern the transcription of hrdB genes in Streptomyces. A cryo-EM structure of Streptomyces coelicolor σShbA-RNAP-promoter open (RPo) complex was solved at 2.97 Å resolution. In combination with in vitro transcription assays, we demonstrate the unique structural features used by the σShbA to recognize the hrdB promoter and form a transcription bubble. All Streptomyces genomes (603) tagged as 'reference' were retrieved from NCBI Datasets. The conserved protein sequences and genomic neighborhoods, as well as the promoter consensus sequences of σShbA and σHrdB homologs, support that the principal σHrdB being governed by the ECF σShbA is a common feature in Streptomyces. Overall, these results provide detailed molecular insights into the transcription of the principal σHrdB gene and pave the way for globally modulating Streptomyces cell viability.
- State Key Laboratory of Microbial Metabolism, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai 200240, China.
Organizational Affiliation: