Structure of beta1-AR-Gs complex bound to epinephrine and an allosteric modulator
Sun, Q., He, G., Xu, X., Liu, X.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Guanine nucleotide-binding protein G(s) subunit alpha isoforms short | 394 | Bos taurus | Mutation(s): 0 Gene Names: GNAS, GNAS1 EC: 3.6.5 | 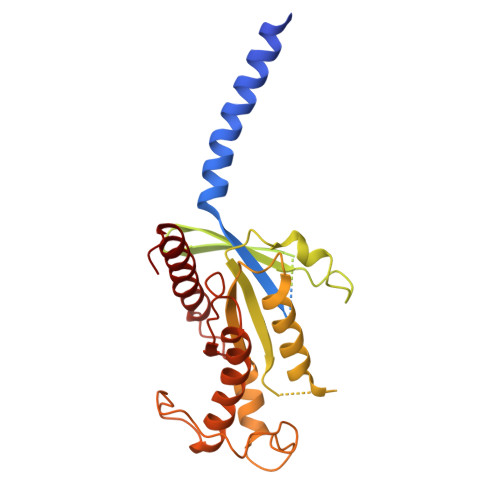 | |
UniProt | |||||
Find proteins for P04896 (Bos taurus) Explore P04896 Go to UniProtKB: P04896 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P04896 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | 351 | Rattus norvegicus | Mutation(s): 0 Gene Names: Gnb1 |  | |
UniProt | |||||
Find proteins for P54311 (Rattus norvegicus) Explore P54311 Go to UniProtKB: P54311 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P54311 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 | C [auth G] | 68 | Bos taurus | Mutation(s): 0 Gene Names: GNG2 | 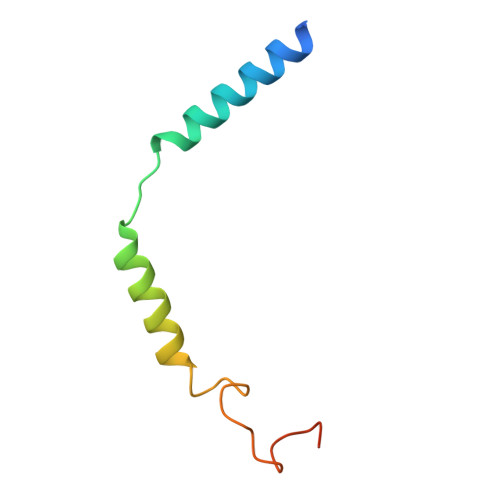 |
UniProt | |||||
Find proteins for P63212 (Bos taurus) Explore P63212 Go to UniProtKB: P63212 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P63212 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| NB35 | D [auth N] | 138 | Lama glama | Mutation(s): 0 | 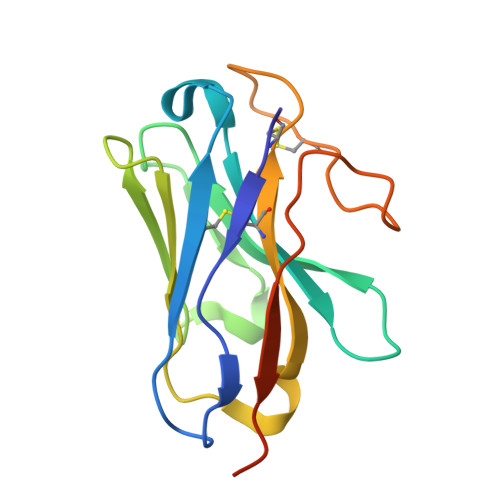 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Beta-1 adrenergic receptor | E [auth R] | 477 | Homo sapiens | Mutation(s): 0 Gene Names: ADRB1, ADRB1R, B1AR | 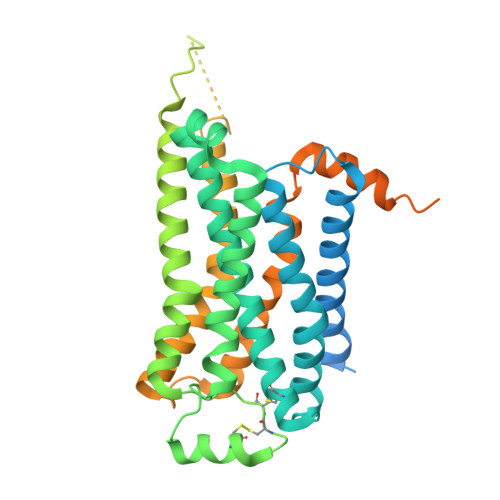 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P08588 (Homo sapiens) Explore P08588 Go to UniProtKB: P08588 | |||||
PHAROS: P08588 GTEx: ENSG00000043591 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P08588 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| DR7 (Subject of Investigation/LOI) Query on DR7 | F [auth R] | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5,
6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER C38 H52 N6 O7 AXRYRYVKAWYZBR-GASGPIRDSA-N |  | ||
| ALE (Subject of Investigation/LOI) Query on ALE | G [auth R] | L-EPINEPHRINE C9 H13 N O3 UCTWMZQNUQWSLP-VIFPVBQESA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | 32122041 |
| National Natural Science Foundation of China (NSFC) | China | 32100968 |