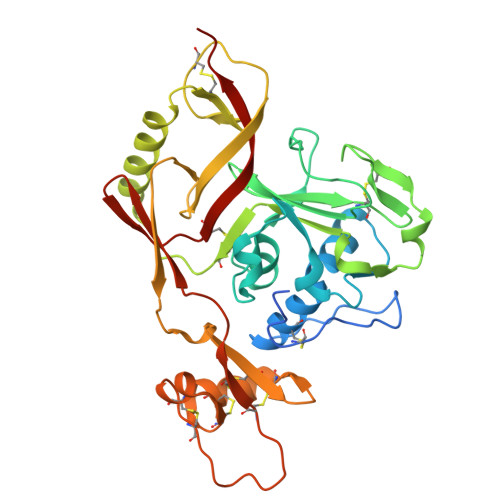
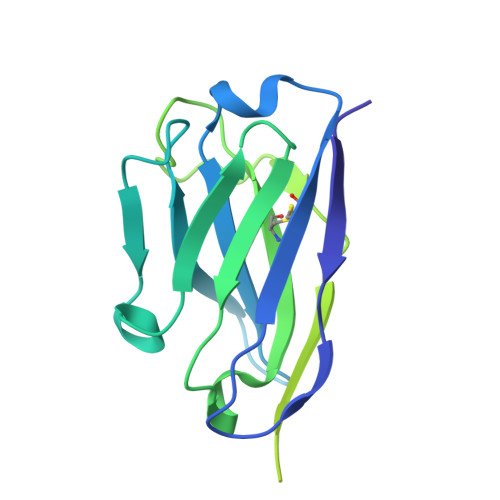
Molecular mechanism of potently neutralizing human monoclonal antibodies against severe fever with thrombocytopenia virus infection.
Quan, C., Nie, K., Ma, D., Su, C., Li, L., Zheng, W., Yin, C., Wang, Y., Yang, P., Peng, D., Liu, X., Li, W., Liu, W., Shan, C., Zheng, J., Liu, D., Zhang, H., Carr, M.J., Gao, G.F., Qi, J., Shi, W.(2025) J Virol 99: e0053325-e0053325
- PubMed: 40539781
- DOI: https://doi.org/10.1128/jvi.00533-25
- Primary Citation of Related Structures:
9JQU, 9JQV, 9L2K - PubMed Abstract:
Although severe fever with thrombocytopenia syndrome (SFTS) was first described in China in 2009, the case fatality rate remains >40% among patients with multi-organ failure. To date, no antivirals specifically targeting SFTSV have been approved. We obtained several monoclonal antibodies (mAbs) from SFTS survivors by single-cell RNA-seq. Neutralization and animal experiments were applied to assess the effects of these mAbs in vitro and in vivo , and co-crystal structures with SFTSV-Gn glycoproteins were determined by X-ray crystallography. The mAbs SD4, SD12, and SD22 targeted the SFTSV-Gn with high neutralizing activities, and, remarkably, SD4 and SD22 exhibited K D values in the range of 32-83 pM for different viral genotypes. Notably, a single administration (20 mg/kg) of SD4 and SD22 showed 100% protection in mice at day 3 post-inoculation (dpi). Importantly, SD4 also provided 60% protection at a lower dose (0.3 mg/kg) when administered at 3 dpi. The crystallographic structures of SD4, SD22, and SD12 with Gn were determined at 3.3 Å, 2.8 Å, and 2.4 Å, respectively, which revealed that they recognized a conserved antigenic epitope around the hexon wellhead edge. These human-derived mAbs have significant therapeutic potential for severe SFTS cases and provide a basis for rational antibody-based vaccine designs and clinical trials. The incidence of severe fever with thrombocytopenia syndrome (SFTS) has been increasing in Asia in recent years; however, no specific antiviral agents have been approved to date. Herein, we report a panel of anti-SFTSV Gn monoclonal antibodies (mAbs) with excellent neutralizing capacities and remarkable therapeutic potential in vitro and in vivo in a mouse model. In addition, crystallographic structures of mAbs complexed with Gn were resolved with atomic resolution (2.4 Å-3.3 Å), revealing a conserved antigenic epitope near the hexon wellhead edge. In sum, the neutralizing antibodies reported in the present study have significant therapeutic potential, paving the way for effective treatment of severe SFTS patients.
- Key Laboratory of Emerging Infectious Diseases in Universities of Shandong, Shandong First Medical University & Shandong Academy of Medical Sciences, Ji'nan, China.
Organizational Affiliation: