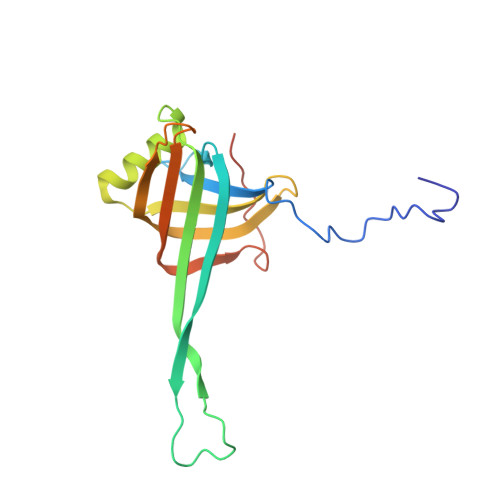
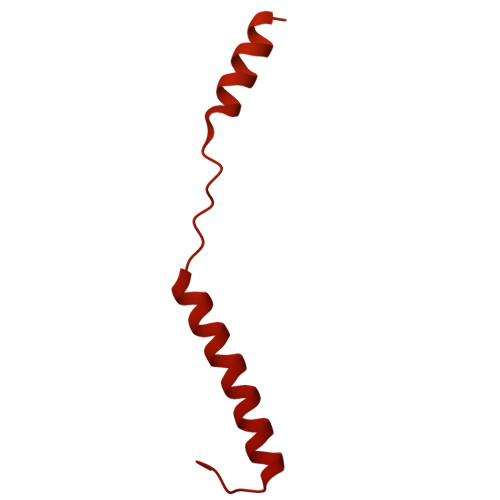
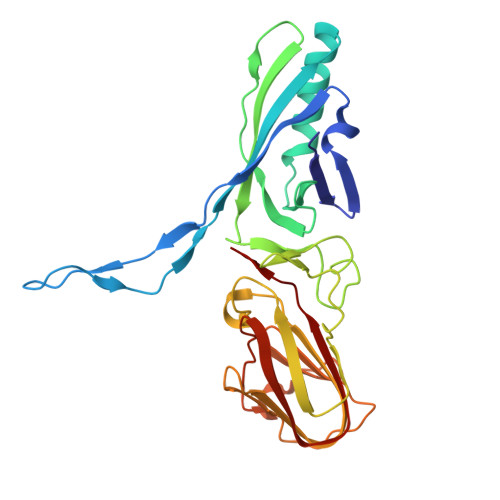
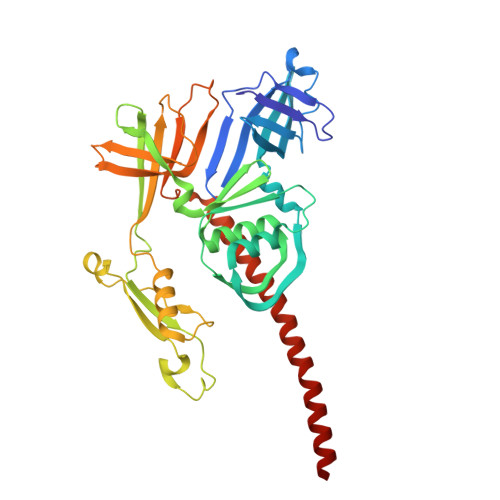
Structure of an F-type phage tail-like bacteriocin from Listeria monocytogenes.
Gu, Z., Ge, X., Wang, J.(2025) Nat Commun 16: 1695-1695
- PubMed: 39956822
- DOI: https://doi.org/10.1038/s41467-025-57075-3
- Primary Citation of Related Structures:
9J1J, 9J1K, 9J1L - PubMed Abstract:
F-type phage tail-like bacteriocins (PTLBs) are high-molecular-weight protein complexes exhibiting bactericidal activity and share evolutionary similarities with the tails of non-contractile siphoviruses. In this study, we present the atomic structure of monocin, a genetically engineered F-type PTLB from Listeria monocytogenes. Our detailed atomic-level analysis, excluding two chaperone proteins, provides crucial insights into the molecular architecture of F-type PTLBs. The core structure of monocin resembles TP901-1-like phage tails, featuring three side fibers with receptor-binding domains that connect to the baseplate for host adhesion. Based on these findings, we propose a potential mechanism by which F-type PTLBs induce cell death, offering a foundation for developing targeted antibacterial therapies.
- State Key Laboratory of Membrane Biology, Beijing Frontier Research Center for Biological Structure, School of Life Sciences, Tsinghua University, Beijing, PR China.
Organizational Affiliation: