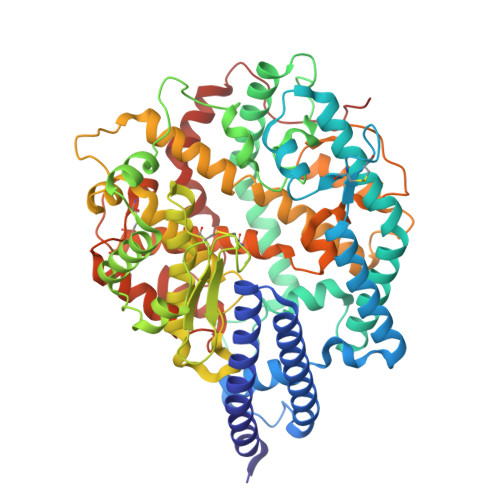
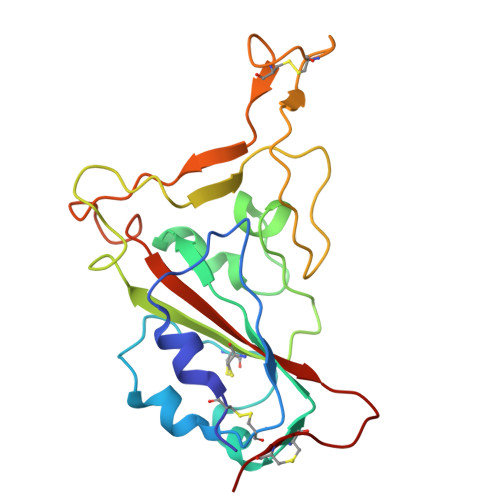
Structural and molecular basis of the epistasis effect in enhanced affinity between SARS-CoV-2 KP.3 and ACE2.
Feng, L., Sun, Z., Zhang, Y., Jian, F., Yang, S., Xia, K., Yu, L., Wang, J., Shao, F., Wang, X., Cao, Y.(2024) Cell Discov 10: 123-123
- PubMed: 39616168
- DOI: https://doi.org/10.1038/s41421-024-00752-2
- Primary Citation of Related Structures:
9IUP, 9IUQ, 9IUU - CAS Key Laboratory of Infection and Immunity, National Laboratory of Macromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China.
Organizational Affiliation: