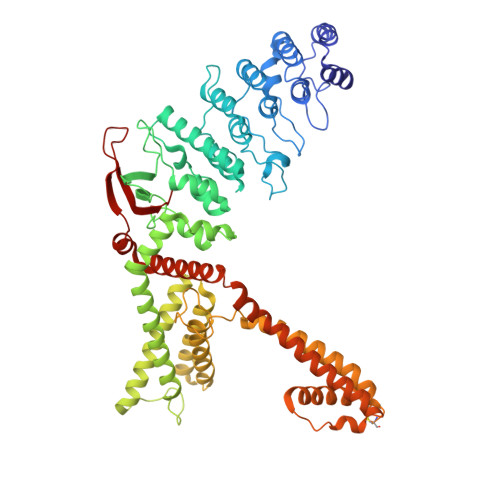
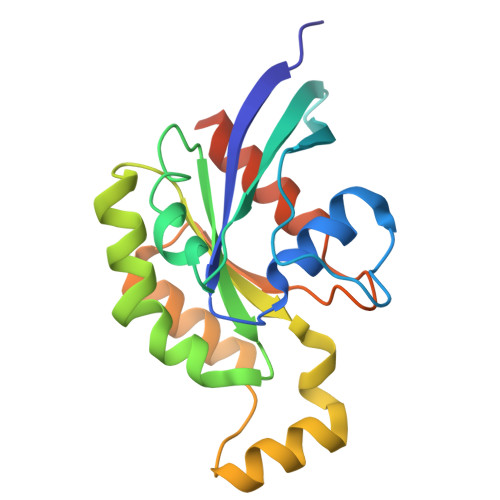
Inactivation of RhoA for Hypertension Treatment Through the TRPV4-RhoA-RhoGDI1 Axis.
Wang, J., Yuan, Z., Yu, N., Jiao, Q., Zhou, H., Liao, W., Shan, J., Ruan, S., Zhao, Y., Mo, Y., Qi, L., Li, T., Fu, J., Ke, B., Xu, Y., Qian, X., Zhang, J., Zhao, Z., Li, S., Wang, R., Li, H.(2025) Circulation 152: 519-536
- PubMed: 40518994
- DOI: https://doi.org/10.1161/CIRCULATIONAHA.124.071884
- Primary Citation of Related Structures:
9IQX, 9IQY - PubMed Abstract:
The RhoA (Ras homolog family member A) signaling pathway is pivotal in regulating vascular smooth muscle cells (VSMCs) function and blood pressure homeostasis. Current inhibitors of the RhoA signaling pathway are limited in hypertension treatment, suffering from poor efficacy, insufficient specificity, and developmental challenges. Cryo-electron microscopy (EM), proximity ligation assay (PLA), and site-directed mutagenesis were used to explore the mechanism of RhoA activity regulation. VSMC, hypertensive animal models, Trpv4 -/- and Arhgdia f/f Myh11-CRE ERT2 (smooth muscle-specific RhoGDI1 knockout) mice were used to investigate the role of the TRPV4 (transient receptor potential cation channel subfamily V member 4)-RhoA-RhoGDI1 (Rho GDP dissociation inhibitor 1) axis in hypertension. AH001 ((R)-1-(3-ethylphenyl) ethane-1,2-diol) was identified as a novel inhibitor of the RhoA signaling pathway. It targets the TRPV4-RhoA-RhoGDI1 axis to effectively sequester inactive RhoA-GDP in the plasma membrane and cytoplasm, which is distinct from typical RhoA inhibition modes. The cryo-EM structure of the TRPV4 AH001 -RhoA complex showed that AH001-bound TRPV4 adopts a closed state with RhoA in an inactive GDP-bound state. Functional studies further revealed that AH001 reduced the pool of active RhoA by enhancing TRPV4-RhoA binding and facilitating RhoGDI1-RhoA interaction in VSMC. This inhibition notably decreased both acute and long-term blood pressure and prevented vascular remodeling in Ang II-induced hypertensive mice and spontaneously hypertensive rats. However, these antihypertensive effects were weakened in Trpv4 -/- and Arhgdia f/f Myh11-CRE ERT2 mice. Additionally, AH001 effectively inhibited VSMC contraction via the RhoA/ROCK (Rho-associated protein kinase)/MYPT1 (myosin phosphatase target subunit 1)/MLC (myosin light chain 2) signaling pathway and suppressed VSMC phenotype switching to myofibroblasts through the RhoA/ROCK/LIMK1 (LIM domain kinase)/cofilin/MRTF-A (myocardin-related transcription factor A)/SRF (serum response factor) signaling cascade. TRPV4 and RhoGDI1 knockdown attenuated AH001's inhibition of VSMC contraction and phenotypic switching to myofibroblasts. This study revealed a novel mode of RhoA signaling inhibition targeting the TRPV4-RhoA-RhoGDI1 axis, offering new insights for future antihypertensive drug development and proposing innovative strategies for targeting challenging Rho GTPases.
- Shanghai Key Laboratory of New Drug Design, School of Pharmacy, East China Normal University, Shanghai, China. (J.W., Z.Y., N.Y., Q.J., H.Z., W.L., J.S., S.R., Y.M., L.Q., J.F., Y.X., X.Q., J.Z., Z.Z., S.L., R.W., H.L.).
Organizational Affiliation: