Receptor-substrate competition for the TonB homologue FusB suggests a model for ferredoxin import in Pectobacterium spp
Wojnowska, M., Flores, V., Yelland, T., Fisher, S.R., Bogucka, A., Stott, K., Walker, D.(2025) bioRxiv
Experimental Data Snapshot
Starting Models: experimental
View more details
wwPDB Validation 3D Report Full Report
(2025) bioRxiv
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein TonB,Ferredoxin-1, chloroplastic | 198 | Pectobacterium carotovorum | Mutation(s): 0 Gene Names: ECA0877, PETF | 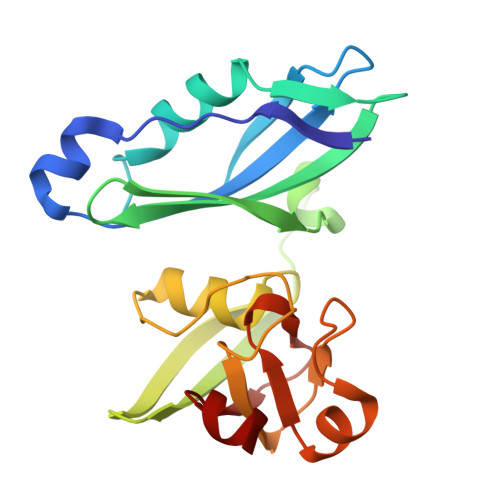 | |
UniProt | |||||
Find proteins for P00221 (Spinacia oleracea) Explore P00221 Go to UniProtKB: P00221 | |||||
Find proteins for Q6D8U5 (Pectobacterium atrosepticum (strain SCRI 1043 / ATCC BAA-672)) Explore Q6D8U5 Go to UniProtKB: Q6D8U5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Groups | Q6D8U5P00221 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| FES Query on FES | B [auth A] | FE2/S2 (INORGANIC) CLUSTER Fe2 S2 NIXDOXVAJZFRNF-UHFFFAOYSA-N |  | ||
| NI Query on NI | C [auth A] | NICKEL (II) ION Ni VEQPNABPJHWNSG-UHFFFAOYSA-N |  | ||
| K Query on K | D [auth A] | POTASSIUM ION K NPYPAHLBTDXSSS-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 66.678 | α = 90 |
| b = 67.446 | β = 90 |
| c = 85.969 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| xia2 | data reduction |
| xia2 | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| University of Cambridge | United Kingdom | -- |