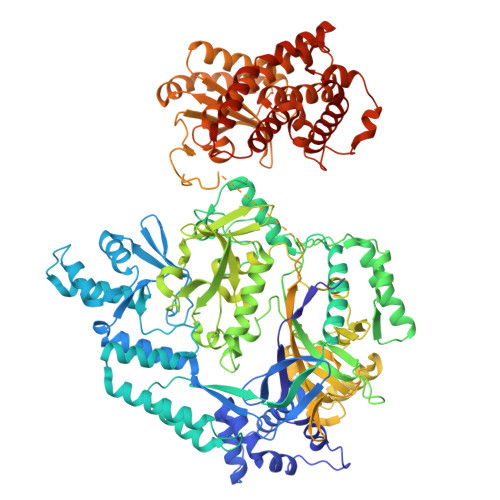
Structural insights into the substrate binding mechanism of the class I dehydratase MadB.
Knospe, C.V., Ortiz, J., Reiners, J., Kedrov, A., Gertzen, C.G.W., Smits, S.H.J., Schmitt, L.(2025) Commun Biol 8: 1032-1032
- PubMed: 40634635
- DOI: https://doi.org/10.1038/s42003-025-08454-5
- Primary Citation of Related Structures:
9G04, 9G05 - PubMed Abstract:
In the battle against antimicrobial resistance, lantibiotics have emerged as promising new sources for antimicrobial drugs. Their exceptional stability is due to characteristic modifications termed (methyl-)lanthionine rings. Genome mining efforts have identified hundreds of lantibiotics by detecting gene operons, so-called biosynthetic gene clusters (BGC), which encode cysteine-rich peptides (30-50 amino acids in size) and enzymes responsible for dehydration and cyclization, catalyzing the post-translational ring formation. One such identified, class I lantibiotic is maddinglicin from Clostridium maddingley. Here, we present single particle cryo-EM structures of the dehydratase MadB in both, its apo-state and in complex with a leader peptide of maddinglicin, revealing a distinct conformational change upon substrate binding. Small-angle X-ray scattering studies elucidate the substrate binding site for the C-terminal part of maddinglicin. Furthermore, a substrate specificity analysis was performed highlighting a critical stretch of amino acids within the maddinglicin leader sequence that is crucial for binding. Here, we provide molecular insights into the conformational changes, principles of substrate recognition and ligand:protein stoichiometry of a class I lantibiotic dehydratase.
- Institute of Biochemistry, Heinrich Heine University Düsseldorf, Düsseldorf, Germany.
Organizational Affiliation: