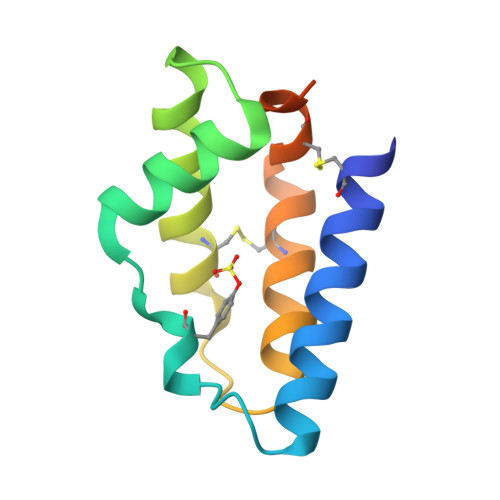
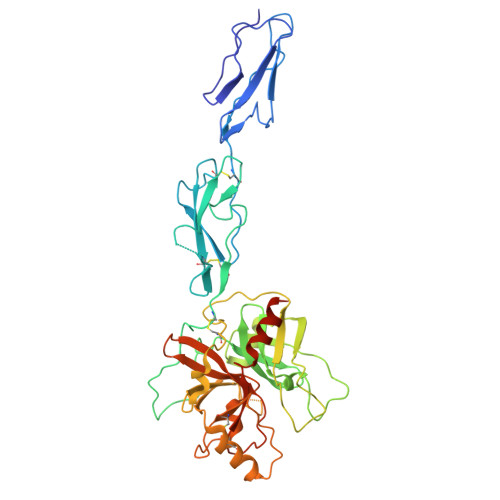
Blocking activation of the C1r zymogen defines a novel mode of complement inhibition.
Duan, H., Wu, W., Li, P., Bouyain, S., Garcia, B.L., Geisbrecht, B.V.(2025) J Biological Chem 301: 108301-108301
- PubMed: 39947467
- DOI: https://doi.org/10.1016/j.jbc.2025.108301
- Primary Citation of Related Structures:
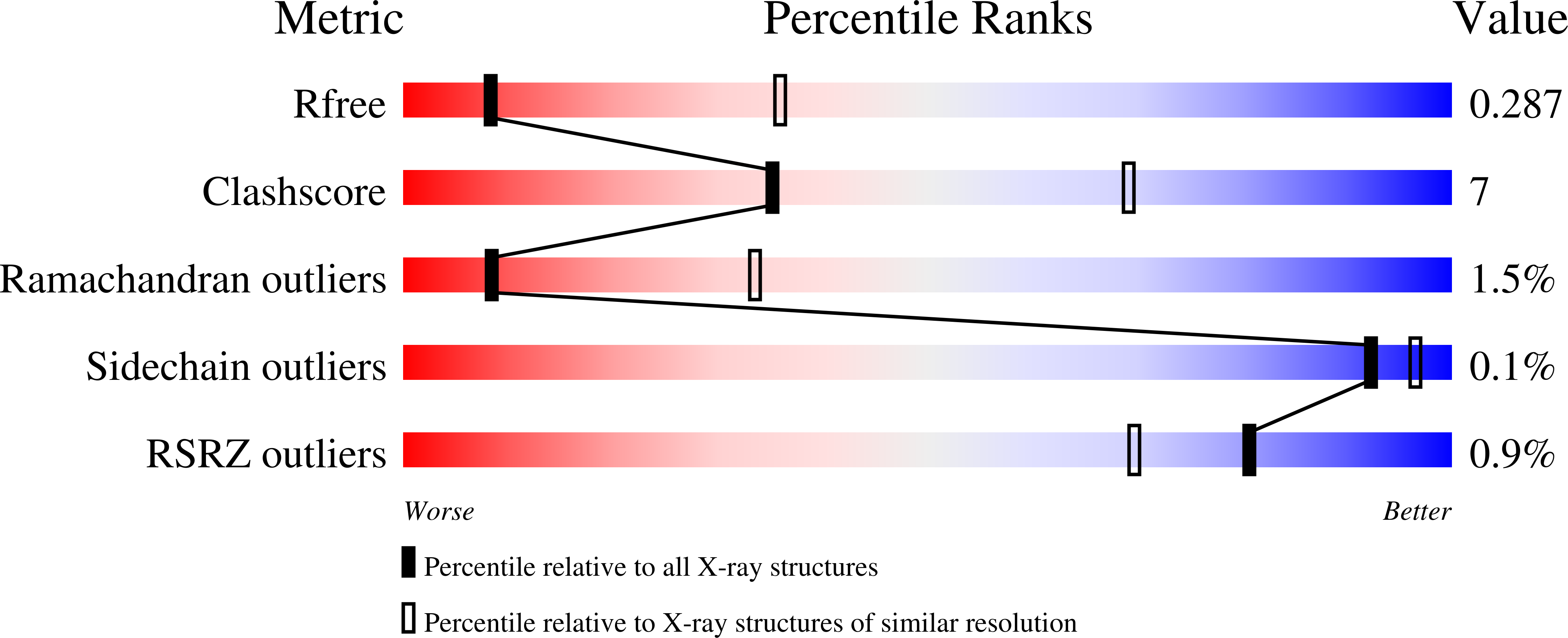
9EKD, 9EKE - PubMed Abstract:
Many hematophagous organisms secrete inhibitors of the coagulation and complement systems as constituents of their salivary fluid. Whereas previous studies on salivary gland extracts from the sandfly Lutzomyia longipalpis identified SALO (salivary anticomplement from L. longipalpis) as a potent inhibitor of the classical complement pathway (CP), its precise mechanism of action has remained elusive. Here, we show that SALO inhibits the CP by binding selectively to the C1r zymogen. Using surface plasmon resonance, we found that SALO expressed by human embryonic kidney 293(T) cells (eSALO-WT) bound with nanomolar affinity to the zymogen of complement protease C1r (pro-C1r), but that it did not bind the enzymatically active form of C1r. To gain insight into the structural basis for CP inhibition by SALO, we solved a 3.3 Å resolution crystal structure of eSALO-WT bound to a recombinant form of C1r that was engineered to remain in a zymogen-like state (zC1r-12SP). eSALO-WT formed extensive interactions with the zymogen activation loop of zC1r-12SP, including groups derived from residues R463 and I464, which compose its scissile peptide bond. Although the interactions with R463 and I464 were mediated by side-chain sulfation of eSALO-WT at position Y51, we found that this modification enhanced the potency of SALO but was not required for its activity. Consistent with our structural observations, subsequent studies showed that eSALO-WT binding to pro-C1r blocked its activation and thereby inhibited the CP in hemolytic assays of complement function. Together, our results define a new mode of inhibiting complement by blocking the farthest upstream enzymatic reaction of the CP.
- Department of Biochemistry & Molecular Biophysics, Kansas State University, Manhattan, Kansas, USA.
Organizational Affiliation: