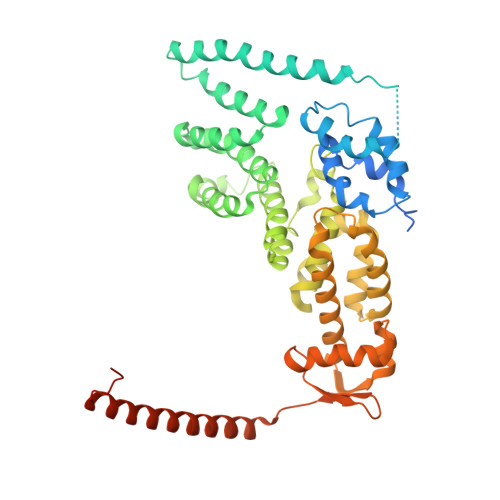
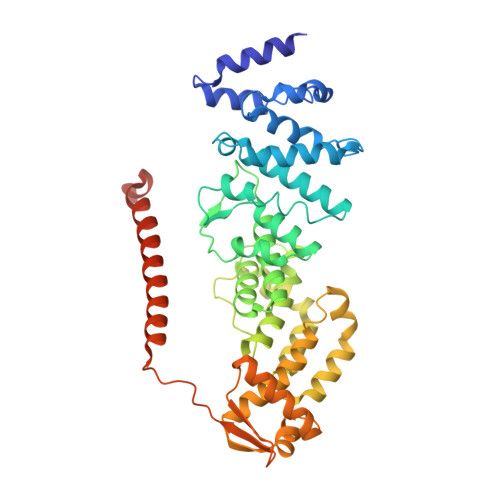
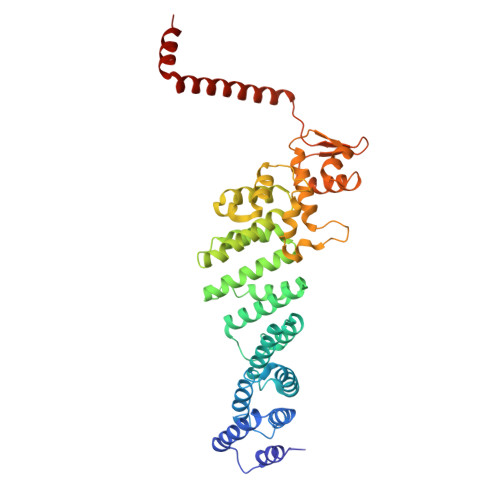
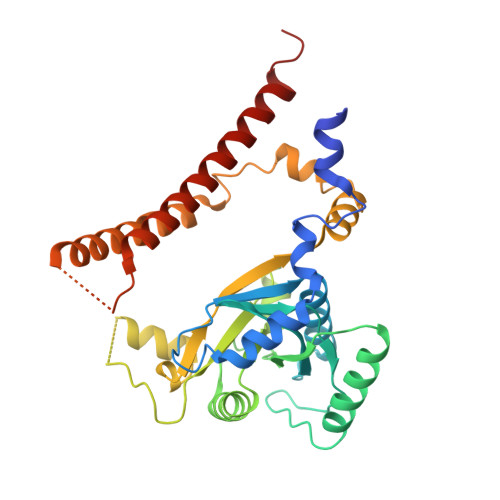
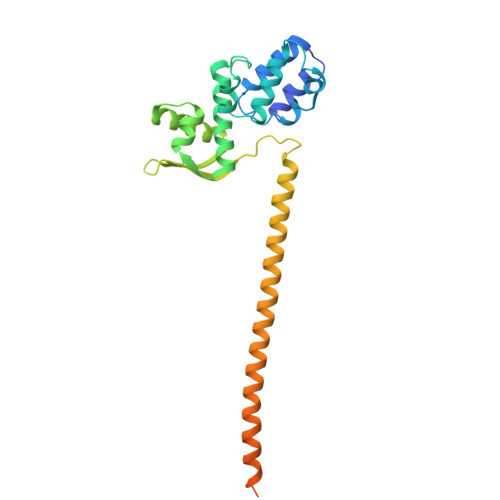
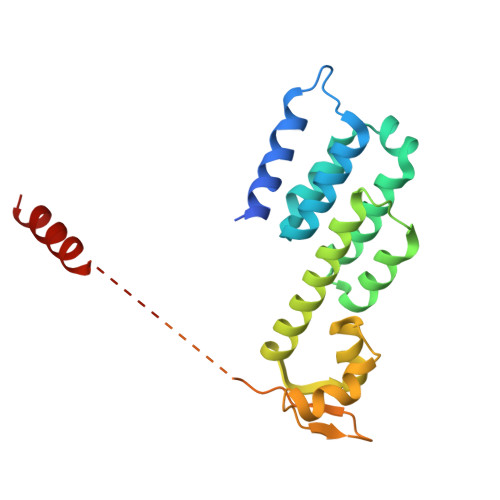
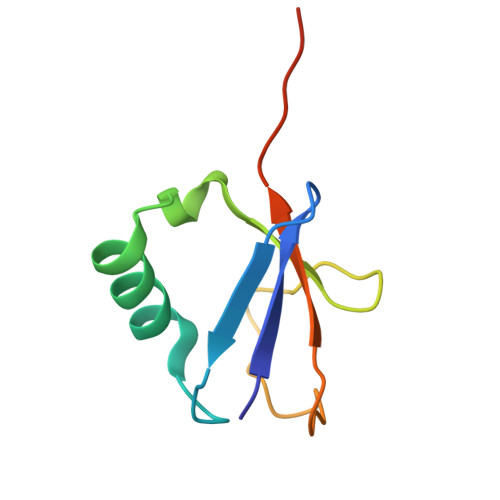
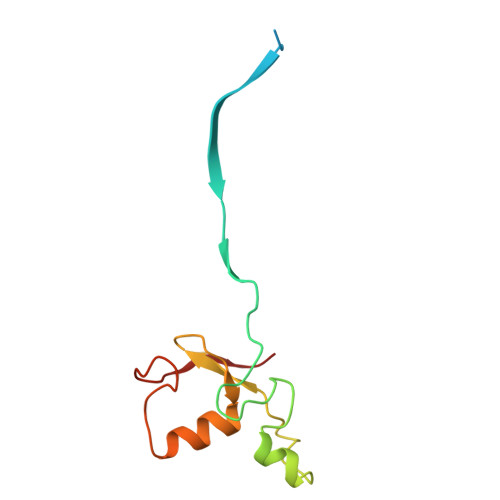
CSN5i-3 is an orthosteric molecular glue inhibitor of COP9 signalosome.
Shi, H., Wang, X., Yu, C., Mao, H., Jiao, F., Braitbard, M., Shor, B., Zhang, Z., Hinds, T.R., Cao, S., Fan, E., Schneidman-Duhovny, D., Huang, L., Zheng, N.(2026) Nature
- PubMed: 41673158
- DOI: https://doi.org/10.1038/s41586-026-10129-y
- Primary Citation of Related Structures:
9E5Z, 9E77, 9E81, 9EFM, 9EFQ, 9EFV, 9EG1, 9EG8, 9EGL, 9PH4 - PubMed Abstract:
Orthosteric inhibitors block enzyme active sites and prevent substrates from binding 1 . Enhancing their specificity through substrate dependence seems inherently unlikely, as their mechanism hinges on direct competition rather than selective recognition. Here we show that a molecular glue mechanism unexpectedly imparts substrate-dependent potency to CSN5i-3, an orthosteric inhibitor of the COP9 signalosome (CSN). We first confirm that CSN5i-3 inhibits CSN, which catalyses NEDD8 (N8) deconjugation from the cullin-RING ubiquitin ligases, by occupying the active site of its catalytic subunit, CSN5, and directly competing with the iso-peptide bond substrate. Notably, the orthosteric inhibitor binds free CSN with only micromolar affinity, yet achieves nanomolar potency in blocking its deneddylase activity. Cryogenic electron microscopy structures of the enzyme-substrate-inhibitor complex reveal that active site-engaged CSN5i-3 occludes the substrate iso-peptide linkage while simultaneously extending an N8-binding exosite of CSN5, acting as a molecular glue to cement the N8-CSN5 interaction. The cooperativity of this trimolecular CSN5i-3-N8-CSN5 assembly, in turn, sequesters CSN5i-3 at its binding site, conferring high potency to the orthosteric inhibitor despite its low affinity for the free enzyme. Together, our findings highlight the modest affinity requirements of molecule glues for individual target proteins and establish orthosteric molecular glue inhibitors as a new class of substrate-dependent enzyme antagonists.
- Department of Pharmacology, University of Washington, Seattle, WA, USA.
Organizational Affiliation: