Characterization of the OMP biogenesis machinery in Fusobacterium nucleatum.
Cottom, C.O., Heinz, E., Erramilli, S., Kossiakoff, A., Slade, D.J., Noinaj, N.(2025) Structure 33: 1878
- PubMed: 40897170
- DOI: https://doi.org/10.1016/j.str.2025.08.008
- Primary Citation of Related Structures:
9CCG - PubMed Abstract:
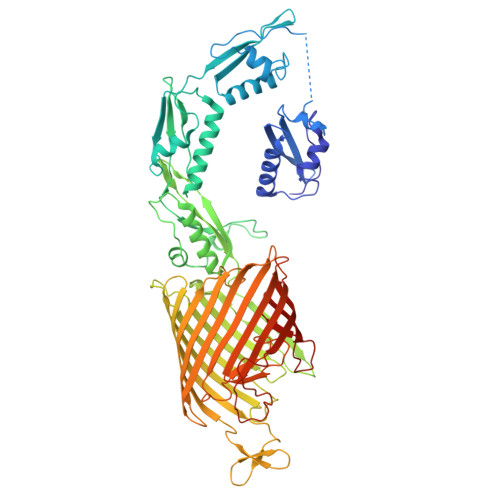
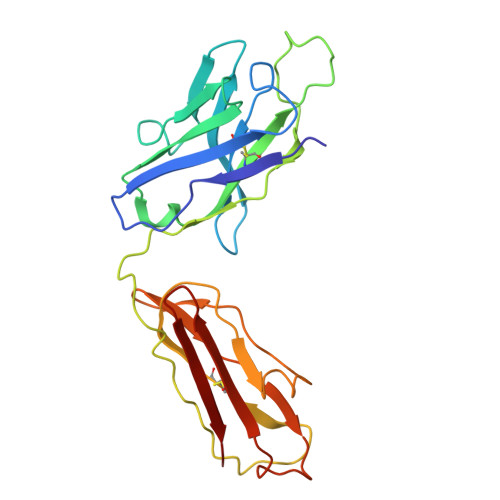
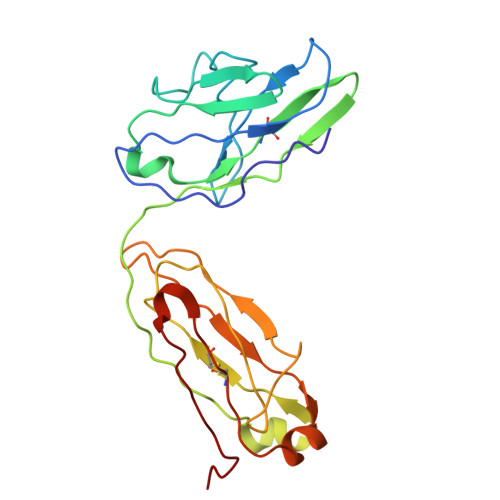
F. nucleatum is a Gram-negative bacteria that causes oral infections and is linked to colorectal cancer. Pathogenicity relies on a type of β-barrel outer membrane protein (OMP) called an autotransporter. The biogenesis of OMPs is typically mediated by the barrel assembly machinery (BAM) complex. In this study, we investigate the evolution, composition, and structure of the OMP biogenesis machinery in F. nucleatum. Our bioinformatics and proteomics analyses indicate that OMP biogenesis in F. nucleatum is mediated solely by the core component BamA. The structure of FnBamA highlights distinct features, including four POTRA domains and a C-terminal 16-stranded β-barrel domain observed as an inverted dimer. FnBamA represents the original composition of the assembly machinery, and a duplication event that resulted in BamA and TamA occurred after the split of other lineages, including the Proteobacteria, from the Fusobacteria. FnBamA, therefore, likely serves a singular role in the biogenesis of all OMPs.
- Department of Biological Sciences, Purdue University, West Lafayette, IN, USA.
Organizational Affiliation: