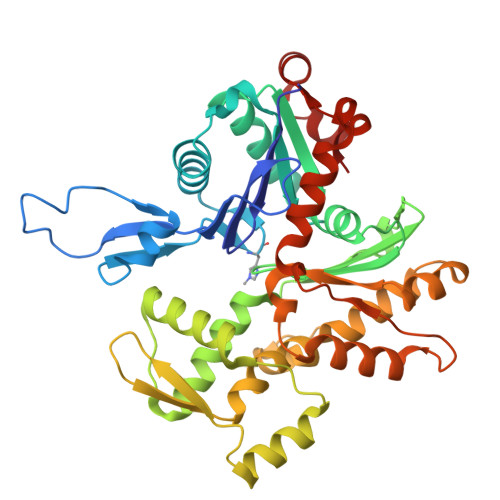
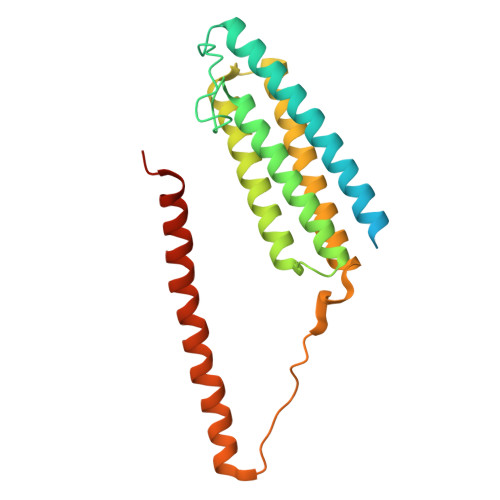
Biochemical and structural bases for talin ABSs-F-actin interactions.
Biertumpfel, C., Yamada, Y., Vasquez-Montes, V., Truong, T.V., Cada, A.K., Mizuno, N.(2025) Proc Natl Acad Sci U S A 122: e2405922122-e2405922122
- PubMed: 39903122
- DOI: https://doi.org/10.1073/pnas.2405922122
- Primary Citation of Related Structures:
9AZ6 - PubMed Abstract:
Focal adhesions (FAs) are large intracellular macromolecular assemblies that play a critical role in cell polarization and migration. Talin serves as a direct connection between integrin receptor and actomyosin cytoskeleton within FAs. Talin contains three actin-binding sites (ABS1-3) that engage discreetly during the development of FAs, thus acting as a critical player in FA initiation and maturation. However, the molecular basis of the ABS-F-actin interactions remains unknown. Here, we explore interactions of ABSs with F-actin to understand the multivalent behavior of talin. Particularly, the cryo-EM structure of the F-actin-ABS3 complex at 2.9 Å shows ABS3 spanning through two actin monomers along the filament axis, each occupied by the R13 rod subdomain and the DD domain. The dimerization of ABS3 occurs through the DD domain where both protomers interact on the actin surface, and the dimerization of talin to the actin surface is necessary for the engagement to F-actin. The R13 helical bundle is distorted upon binding to F-actin and releases the H1 helix from the rest of the bundle. This phenomenon has also been observed with other tension-sensing proteins like vinculin and α-catenin, highlighting that unfolding is relevant for its force sensing activity. On the contrary, ABS2 (R4R8 subdomains), which is thought to be critical for the maintenance of mature FAs, had multiple F-actin-binding regions within ABS2 and the binding likely occurred by these subdomains running through the surface of F-actin, thus strengthening the interactions upon the maturation of FAs.
- Laboratory of Structural Cell Biology, National Heart, Lung, and Blood Institute, NIH, Bethesda, MD 20892.
Organizational Affiliation: