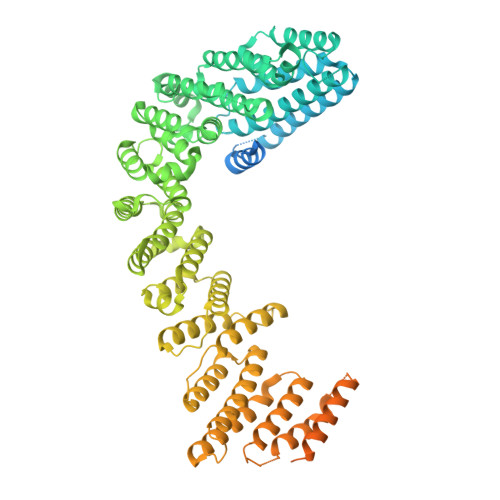
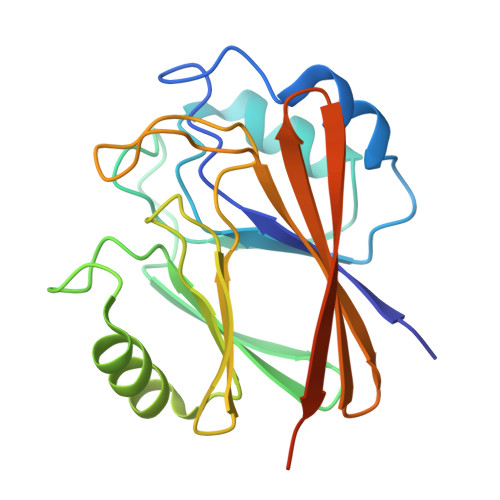
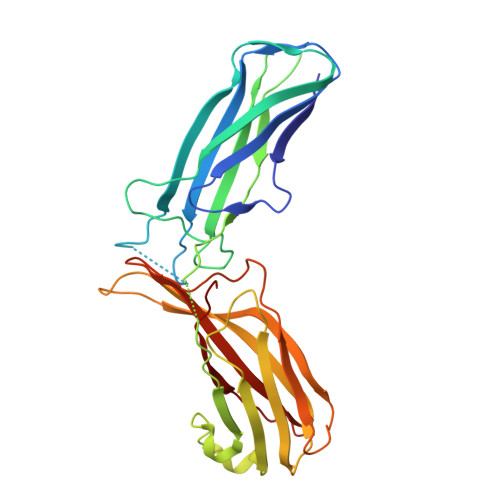
Structural basis for Retriever-SNX17 assembly and endosomal sorting.
Singla, A., Boesch, D.J., Fung, H.Y.J., Ngoka, C., Enriquez, A.S., Song, R., Kramer, D.A., Han, Y., Banarer, E., Lemoff, A., Juneja, P., Billadeau, D.D., Bai, X., Chen, Z., Turer, E.E., Burstein, E., Chen, B.(2024) Nat Commun 15: 10193-10193
- PubMed: 39587083
- DOI: https://doi.org/10.1038/s41467-024-54583-6
- Primary Citation of Related Structures:
9AU7 - PubMed Abstract:
During endosomal recycling, Sorting Nexin 17 (SNX17) facilitates the transport of numerous membrane cargo proteins by tethering them to the Retriever complex. Despite its importance, the mechanisms underlying this interaction have remained elusive. Here, we provide biochemical, structural, cellular, and proteomic analyses of the SNX17-Retriever interaction. Our data reveal that SNX17 adopts an autoinhibited conformation in the basal state, with its FERM domain sequestering its C-terminal tail. The binding of cargo proteins to the FERM domain displaces the C-terminal tail through direct competition. The released tail engages with Retriever by binding to a highly conserved interface between its VPS35L and VPS26C subunits, as revealed by cryogenic electron microscopy (cryo-EM). Disrupting this interface impairs the Retriever-SNX17 interaction, subsequently affecting the recycling of SNX17-dependent cargoes and altering the composition of the plasma membrane proteome. Intriguingly, the SNX17-binding pocket on Retriever can be utilized by other ligands containing a consensus acidic C-terminal tail motif. Together, our findings uncover a mechanism underlying endosomal trafficking of critical cargo proteins and reveal how Retriever can potentially engage with other regulatory factors or be exploited by pathogens.
- Department of Internal Medicine, University of Texas Southwestern Medical Center, 5323 Harry Hines Boulevard, Dallas, TX, 75390, USA.
Organizational Affiliation: