UV lesion binding and repair inhibition by ETS-family transcription factors
Sivapragasam, S., Terrell, J.R., Germann, M.W., Morledge-Hampton, B., Adhikari, S.P., Brown, M., Hrdlicka, P.J., Wyrick, J.J., Poon, G.M.K.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Transcription factor PU.1 | C [auth F] | 106 | Homo sapiens | Mutation(s): 0 Gene Names: SPI1 | 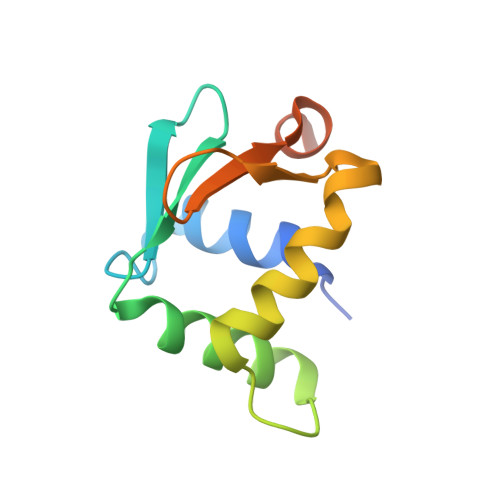 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P17947 (Homo sapiens) Explore P17947 Go to UniProtKB: P17947 | |||||
PHAROS: P17947 GTEx: ENSG00000066336 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P17947 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*AP*AP*TP*AP*AP*GP*CP*GP*GP*AP*AP*GP*TP*GP*GP*G)-3') | A [auth C] | 16 | synthetic construct | 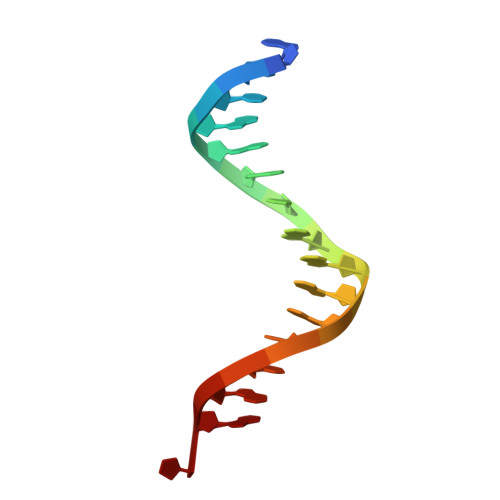 | |
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(P*CP*CP*CP*AP*CP*T*(CPD)P*CP*GP*CP*TP*TP*AP*T)-3') | B [auth D] | 15 | synthetic construct | 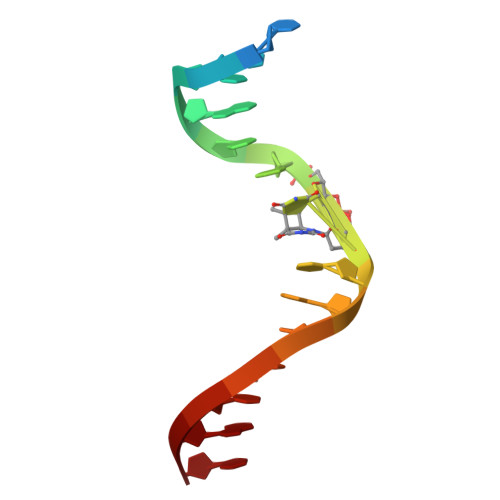 | |
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 42.966 | α = 90 |
| b = 58.879 | β = 117.593 |
| c = 45.925 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of Environmental Health Sciences (NIH/NIEHS) | United States | ES035139 |
| National Institutes of Health/National Heart, Lung, and Blood Institute (NIH/NHLBI) | United States | HL155178 |
| National Science Foundation (NSF, United States) | United States | 2028902 |