Cryo-EM structure of post-fusion EBV gB in complex with AMMO2 fab
Chou, C.W., McCool, R.S., McLellan, J.S.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| AMMO2 fab heavy chain | A [auth F], C [auth H], E [auth D] | 133 | Homo sapiens | Mutation(s): 0 | 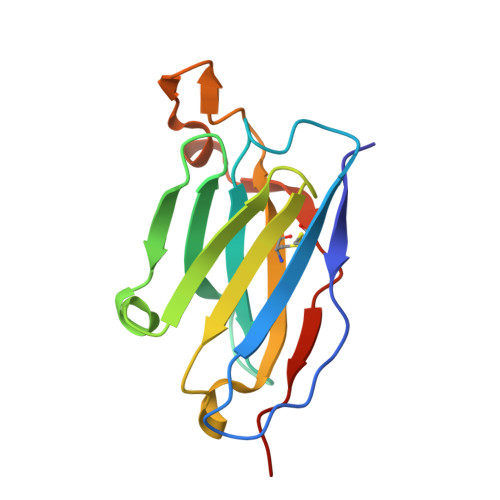 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| AMMO2 fab light chain | B [auth G], D [auth I], F [auth E] | 214 | Homo sapiens | Mutation(s): 0 | 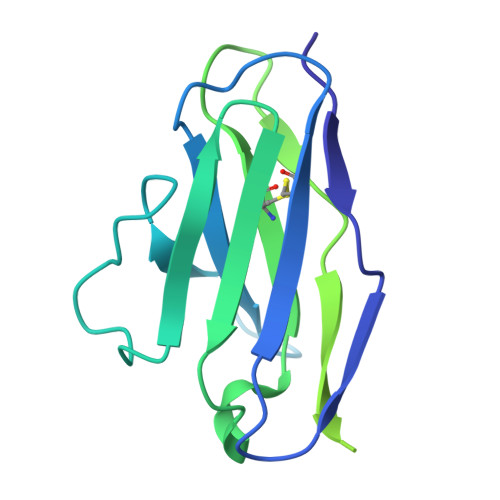 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| BALF4 | G [auth A], H [auth B], I [auth C] | 734 | human gammaherpesvirus 4 | Mutation(s): 0 Gene Names: BALF4 | 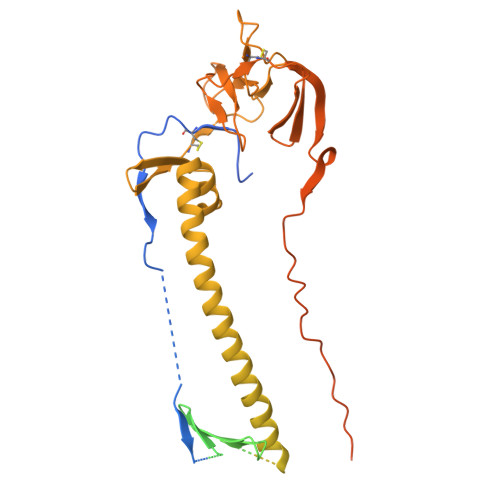 |
UniProt | |||||
Find proteins for P0C762 (Epstein-Barr virus (strain GD1)) Explore P0C762 Go to UniProtKB: P0C762 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0C762 | ||||
Glycosylation | |||||
| Glycosylation Sites: 2 | |||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | J [auth A] K [auth A] L [auth B] M [auth B] N [auth C] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.21.2_5419 |
| RECONSTRUCTION | cryoSPARC |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Other private | United States | -- |