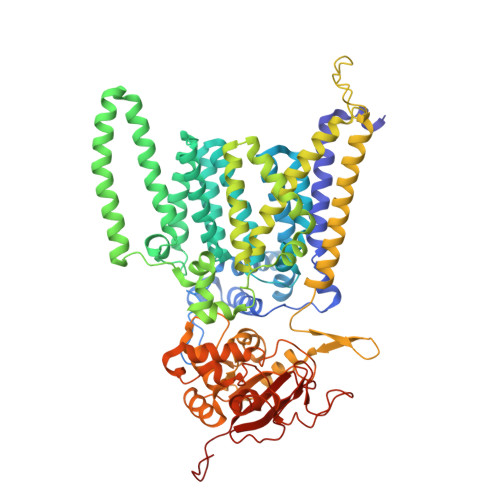
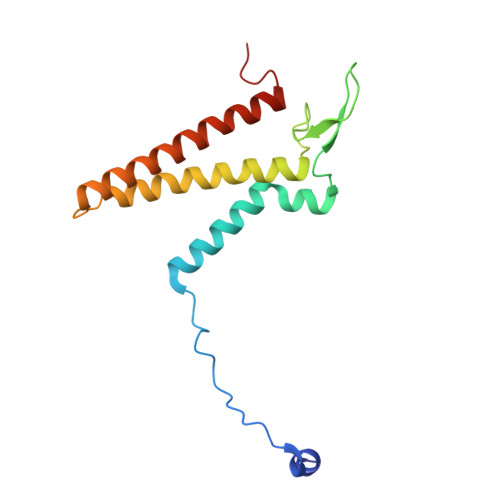
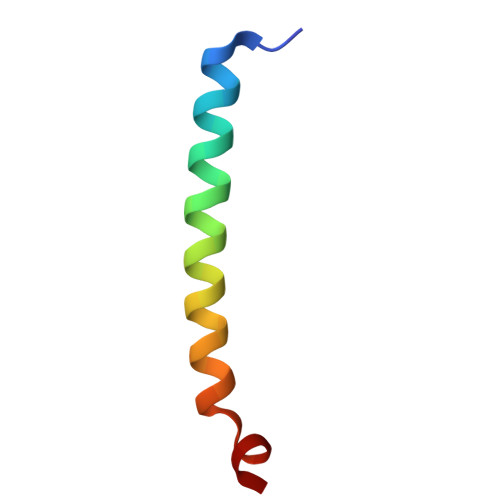
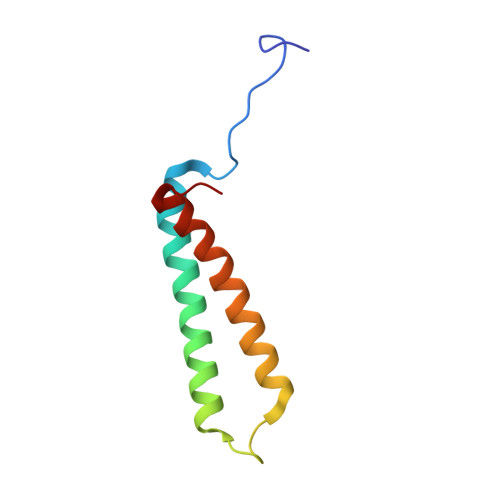
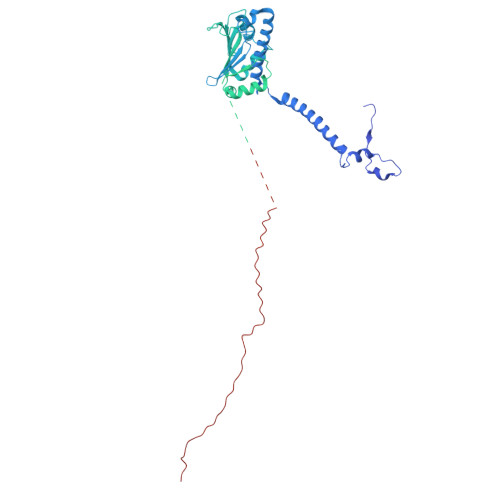
Structural basis of regulated N-glycosylation at the secretory translocon.
Yamsek, M., Ma, M., Jha, R., Wan, Y., Li, Q., Zhong, F., DeLong, K., Ji, Z., Rohatgi, R., Keenan, R.J.(2026) Nature 649: 777-784
- PubMed: 41261126
- DOI: https://doi.org/10.1038/s41586-025-09756-8
- Primary Citation of Related Structures:
9N9J, 9YGY - PubMed Abstract:
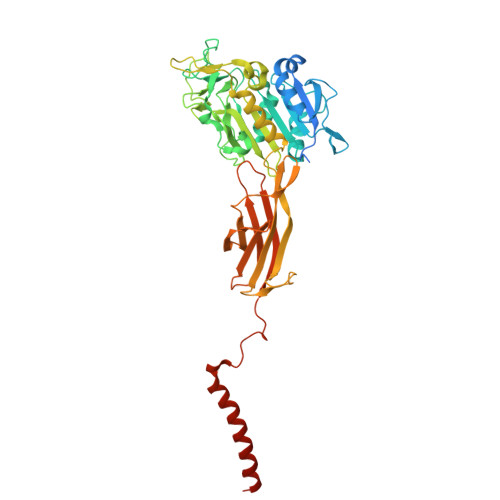
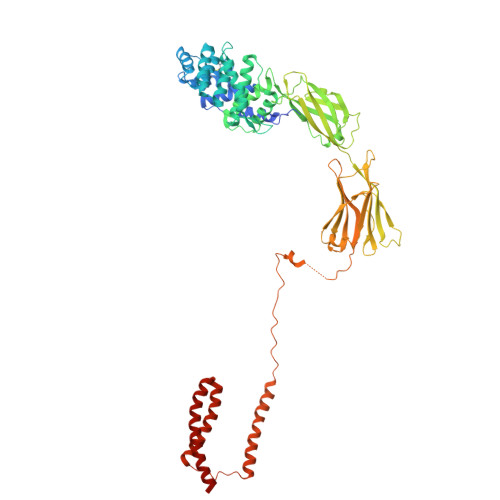
Most human secretory pathway proteins are N-glycosylated by oligosaccharyltransferase (OST) complexes as they enter the endoplasmic reticulum (ER) 1-3 . Recent work revealed a substrate-assisted mechanism by which N-glycosylation of the chaperone glucose-regulated protein 94 (GRP94) is regulated to control cell surface receptor signalling 4 . Here we report the structure of a natively isolated GRP94 folding intermediate tethered to a specialized CCDC134-bound translocon. Together with functional analysis, the data reveal how a conserved N-terminal extension in GRP94 inhibits OST-A and how structural rearrangements within the translocon shield the tethered nascent chain from inappropriate OST-B glycosylation. These interactions depend on a hydrophobic CCDC134 groove, which recognizes a non-native conformation of nascent GRP94. Our results define a mechanism of regulated N-glycosylation and illustrate how the nascent chain remodels the translocon to facilitate its own biogenesis.
- Department of Biochemistry and Molecular Biology, The University of Chicago, Chicago, IL, USA.
Organizational Affiliation: