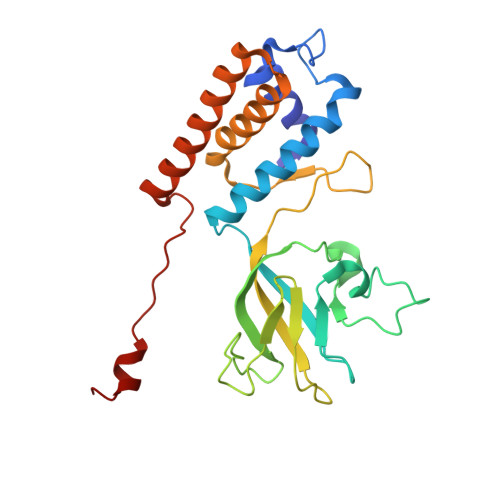
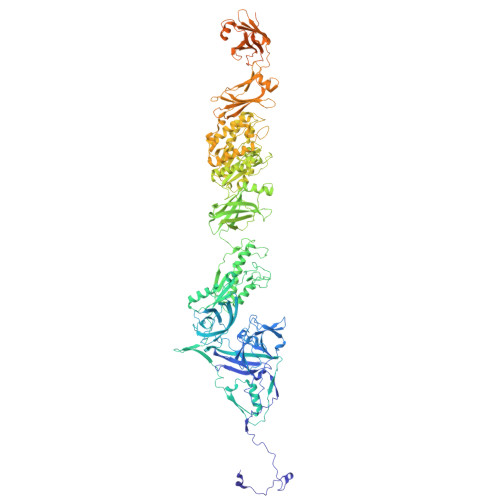
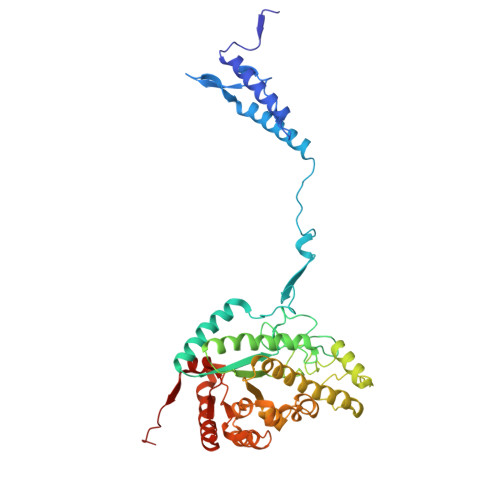
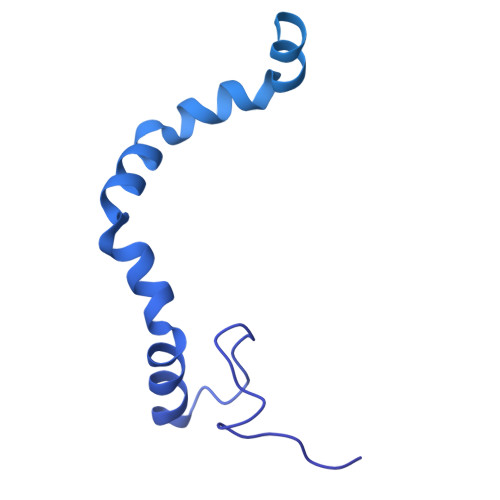
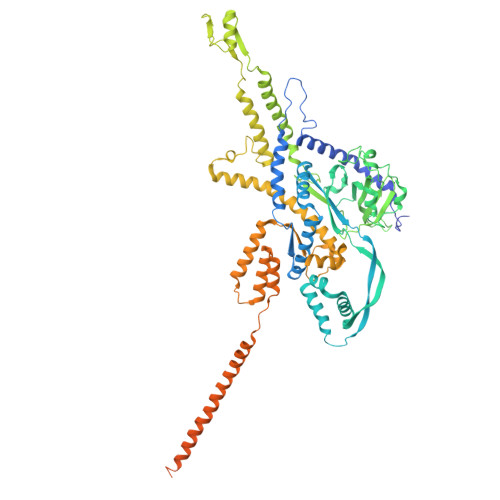
Structure of the giant RNA polymerase ejected from coliphage N4.
Bellis, N.F., Lokareddy, R.K., Pavlenok, M., Horton, S.L.C., Kizziah, J.L., Forti, F., Schneider, D.A., Niederweis, M., Briani, F., Cingolani, G.(2025) Res Sq
- PubMed: 41282253
- DOI: https://doi.org/10.21203/rs.3.rs-7746245/v1
- Primary Citation of Related Structures:
9PNQ, 9PNR, 9PNT, 9PNV, 9PNW, 9YF5, 9YF8 - PubMed Abstract:
Schitoviruses are widespread prokaryotic viruses that encapsidate a giant (~3,500-residue) virion-associated RNA polymerase (vRNAP). During infection, vRNAP is expelled into Gram-negative bacteria, along with two additional ejection proteins, to assemble a transient DNA-ejectosome that becomes transcriptionally active, initiating viral replication. Here, we present an integrative structural analysis of the coliphage N4 vRNAP (gp50). We find that this 383 kDa enzyme is a multi-domain, single-chain RNA polymerase, structurally distinct from both compact single-chain RNAPs and large multi-subunit holoenzymes. vRNAP is composed of loosely connected domains and exhibits an intramolecular mode of allosteric regulation through its C-terminal domain. Comparative analysis of intact and genome-released virions identified gp51, which forms an outer-membrane complex, and gp52, which assembles a periplasmic tunnel. These proteins generate heterogeneous pores that facilitate the release of vRNAP. We further uncover a signaling hub in the phage tail, composed of the receptor-binding protein, tail tube, and tail plug, that detects receptor engagement and orchestrates the release of ejection proteins. We propose that the beads-on-a-string architecture of vRNAP enables the translocation of megadalton-scale protein complexes through the ~35 Å channel formed by the tail and ejection proteins. These findings establish N4 as a distinctive model for protein translocation through biological channels.
- Department of Biochemistry and Molecular Genetics, University of Alabama at Birmingham, 1825 University Blvd, Birmingham, AL 35294, USA.
Organizational Affiliation: