Discovery of Zilucoplan: A Complement C5 Inhibitor for Treatment of Anti-Acetylcholine Receptor (AChR) Antibody-Positive Generalized Myasthenia Gravis (gMG).
Ye, P., Hammer, R.P., Wang, Z., Dhamnaskar, K., Hoarty, M., Ma, Z., Tang, G.Q., DeMarco, S.J., Ricardo, A.(2025) J Med Chem 68: 25772-25782
- PubMed: 41379101
- DOI: https://doi.org/10.1021/acs.jmedchem.5c02537
- Primary Citation of Related Structures:
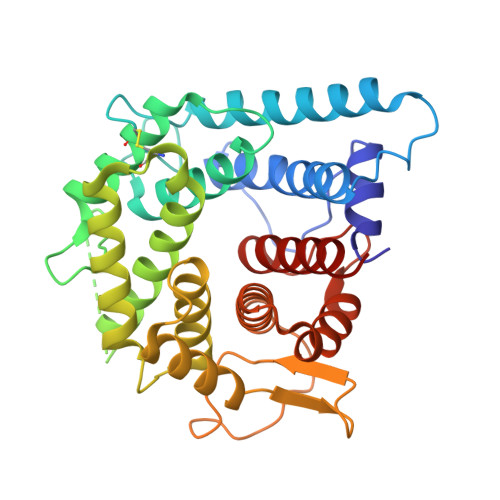
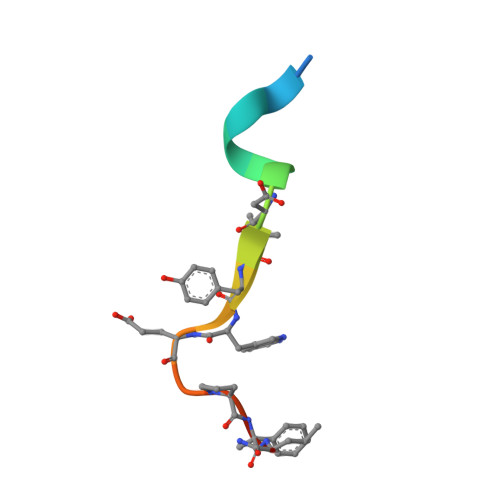
9Y6C - PubMed Abstract:
Complement component 5 (C5) is a protein in the complement cascade and a part of the innate immune system that has been clinically validated as a therapeutic target for several immune-mediated diseases including generalized myasthenia gravis (gMG). In this paper, we discuss the early discovery of zilucoplan, a macrocyclic peptide drug, which was identified via innovative extreme diversity mRNA display (Ma, Z.; Hartman, M. C. T. In Vitro Selection of Unnatural Cyclic Peptide Libraries via mRNA Display. In Ribosome Display and Related Technologies: Methods and Protocols ; Douthwaite, J. A., Jackson, R. H., Eds.; Springer: New York, 2012; pp 367-390.) against C5 and approved for the treatment of gMG. We highlight the key steps and rationale behind the peptide medicinal chemistry optimization of the initial screening hits, that led to improved potency, stability, and pharmacokinetic properties.
- UCB Bioscience, Cambridge, Massachusetts 02140, United States.
Organizational Affiliation: