Targeting mGlyR with nanobodies for depression
Laboute, T., Zucca, S., Sial, O.K., Sharma, M., Brunori, G., Singh, S., Nageswar, K., Peng, H., Rader, C., Becker, J.A., Le Merrer, J., Singh, A.K., Martemyanov, K.A.(2026) Nat Commun 17: 831
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2026) Nat Commun 17: 831
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Metabotropic glycine receptor | 781 | Homo sapiens | Mutation(s): 0 Gene Names: GPR158, KIAA1136 | 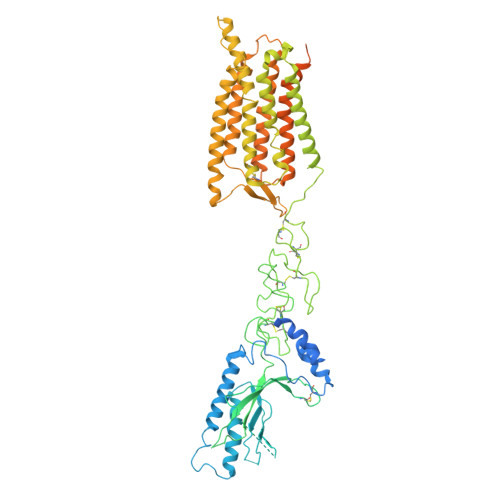 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q5T848 (Homo sapiens) Explore Q5T848 Go to UniProtKB: Q5T848 | |||||
PHAROS: Q5T848 GTEx: ENSG00000151025 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5T848 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Nb20 | C, D [auth E] | 150 | Lama glama | Mutation(s): 0 | 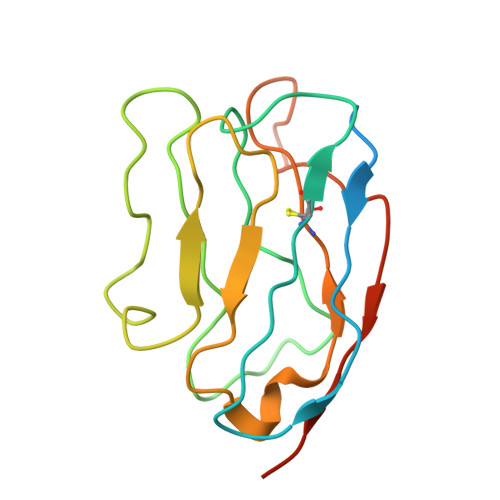 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PIE (Subject of Investigation/LOI) Query on PIE | Z [auth B] | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoinositol C43 H80 O13 P PDLAMJKMOKWLAJ-OJERQSHOSA-M |  | ||
| PEE (Subject of Investigation/LOI) Query on PEE | P [auth A] | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine C41 H78 N O8 P MWRBNPKJOOWZPW-NYVOMTAGSA-N |  | ||
| CLR Query on CLR | E [auth A] F [auth A] G [auth A] H [auth A] I [auth A] | CHOLESTEROL C27 H46 O HVYWMOMLDIMFJA-DPAQBDIFSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cryoSPARC | |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Eye Institute (NIH/NEI) | United States | MH105482 |
| National Institutes of Health/National Eye Institute (NIH/NEI) | United States | EY034339 |
| Indian Council of Medical Research | India | Neuro/2022-NCD-I |
| Department of Biotechnology (DBT, India) | India | Neuro/2022-NCD-I |