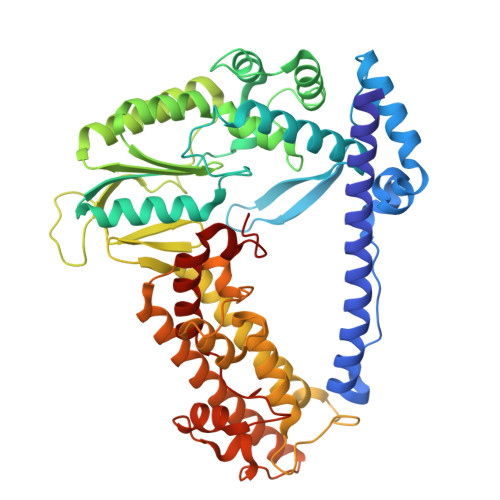
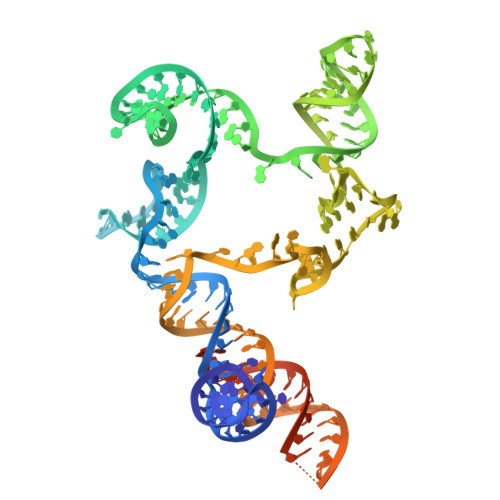
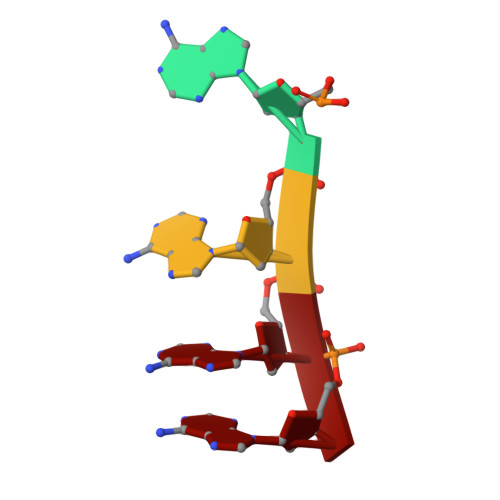
Non-coding RNA mediates the defense-associated reverse transcriptase (DRT) anti-phage oligomerization transition.
Han, J., Liu, B., Tang, J., Zhang, S., Wang, X., Li, X., Zhang, Q., Liu, Z., Wang, W., Liu, Y., Zhou, R., Yin, H., Wei, Y., Li, Z., Zhang, M., Deng, Z., Zhang, H.(2025) EMBO J 44: 5429-5442
- PubMed: 40836036
- DOI: https://doi.org/10.1038/s44318-025-00544-8
- Primary Citation of Related Structures:
9VKU, 9VMA - PubMed Abstract:
Defense-associated reverse transcriptase (DRT) systems are implicated in prokaryotic resistance to viral infections, yet the molecular mechanisms underlying their functionality remain largely unknown. Here, we characterize a two-component DRT9 system, composed of a reverse transcriptase (RT) and a non-coding RNA (ncRNA), which exhibits a protein-primed DNA synthesis activity upon phage infection. We also determine its cryo-electron microscopy (cryo-EM) structures in different functional states. DRT9 RT binds to ncRNA, forming a dimer of dimers configuration that assembles into a trimer of dimers upon substrate binding. This oligomerization transition, crucial for DRT9-mediated anti-phage defense, is facilitated by a ncRNA cooperative self-assembly manner. Furthermore, substrate binding induces large conformational movements around the catalytic pocket of DRT9 RT, revealing a "lock-switch" mechanism for enzymatic activation. Notably, phylogenetic analysis and functional assays identify a unique N-terminal helix extension required for ncRNA stabilization and enzymatic activity, distinct from previously reported reverse transcriptase systems. Overall, our findings illuminate the molecular basis of DRT9-mediated antiviral defense and expand the functional and mechanistic diversity of the DRT family.
- Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), State Key Laboratory of Experimental Hematology, Tianjin Medical University Cancer Institute and Hospital, The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, Tianjin Institute of Immunology, School of Basic Medical Sciences, Tianjin Medical University, Tianjin, China.
Organizational Affiliation: