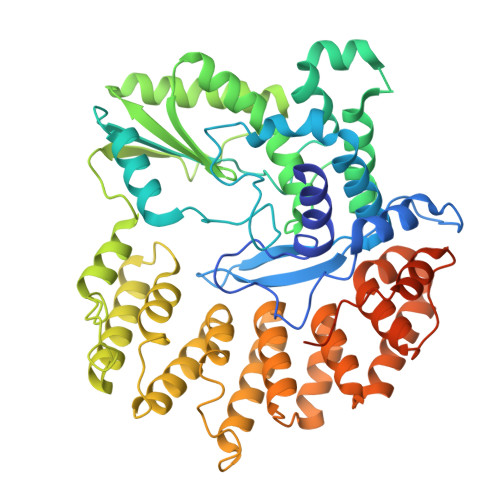
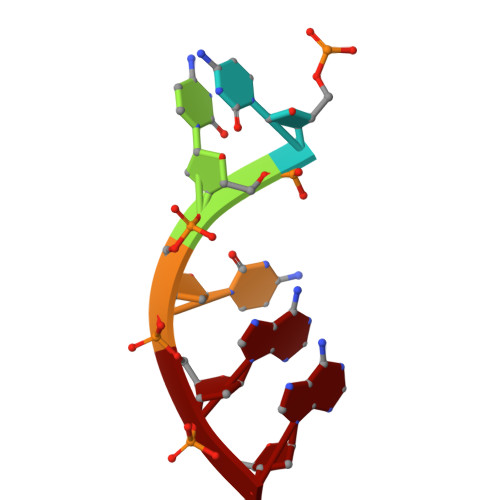
Anti-phage defense mechanism involving phage-encoded DNA binding protein and bacterial reverse transcriptase DRT4.
Wang, Y., Wu, H., Li, J., An, Q., Tian, Z., Deng, Z.(2025) Nat Commun 17: 289-289
- PubMed: 41330908
- DOI: https://doi.org/10.1038/s41467-025-66997-x
- Primary Citation of Related Structures:
9VDO, 9VDP, 9VDV - PubMed Abstract:
Prokaryotic defense-associated reverse transcriptase (DRT) systems confer host resistance to viral infection through DNA synthesis; however, the molecular mechanisms underlying their function remain poorly understood. Here, we demonstrate that DRT4, a single-gene anti-phage defense system, synthesizes single-stranded DNA (ssDNA) products of random sequences in a template-independent manner. High-resolution cryo-EM structures of DRT4 in multiple functional states elucidate its oligomeric architecture, catalytic metal ion coordination, and substrate/DNA product binding, offering mechanistic insights into its promiscuous polymerization activity. Structural and biochemical analyses further identify a conserved tyrosine residue that acts as the priming site for the initiation of DNA synthesis. Upon phage infection, a phage-encoded DNA-binding protein, ORF55, protects the 3' end of the DRT4-synthesized ssDNA from host exonuclease degradation, likely resulting in toxic ssDNA accumulation that leads to cell death. Remarkably, ORF55 also activates DRT6, a structural homolog of DRT4, suggesting a conserved activation mechanism among related DRT systems. These findings provide structural and mechanistic insights into DRT4-mediated antiviral defense, establishing a distinct paradigm for antiviral reverse transcriptase in bacterial immunity.
- State Key Laboratory of Virology and Biosafety, Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan, Hubei, China.
Organizational Affiliation: