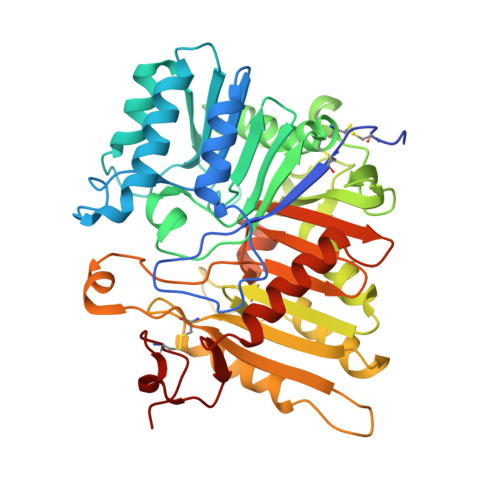
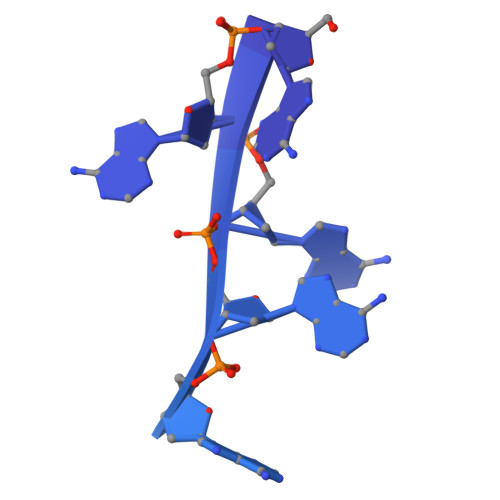
Mechanistic insights into single-stranded DNA degradation by lysosomal exonucleases PLD3 and PLD4 from structural snapshots.
Hirano, Y., Ezaki, W., Sato, R., Ohto, U., Miyake, K., Shimizu, T.(2025) Nat Commun 16: 11431-11431
- PubMed: 41381514
- DOI: https://doi.org/10.1038/s41467-025-66261-2
- Primary Citation of Related Structures:
9VBG, 9VBH, 9VBI, 9VBJ, 9VBK - PubMed Abstract:
Lysosomal exonuclease phospholipase D (PLD) family PLD3 and PLD4 degrade single-stranded RNA or DNA and regulate TLR7 or TLR9 responses. Polymorphisms of these enzymes are associated with human diseases: PLD4 is associated with inflammatory diseases, and PLD3 is associated with neurodegenerative diseases. Here, we determine the structures of substrate-bound PLD3 and PLD4 by cryo-electron microscopy. Our structures reveal that PLD3 rebuilds a substrate-binding pocket, depending on the substrate, mainly via motion of the Phe335-containing loop. Furthermore, we captured the structure in a metastable state that appears during substrate rearrangement following product release. Together, our findings identify the residues that underlie the distinct activities of PLD3 and PLD4. This study provides a mechanistic basis for the exonuclease activity of PLD3 and PLD4 in single-stranded DNA degradation.
- Graduate School of Pharmaceutical Sciences, The University of Tokyo, Tokyo, Japan.
Organizational Affiliation: