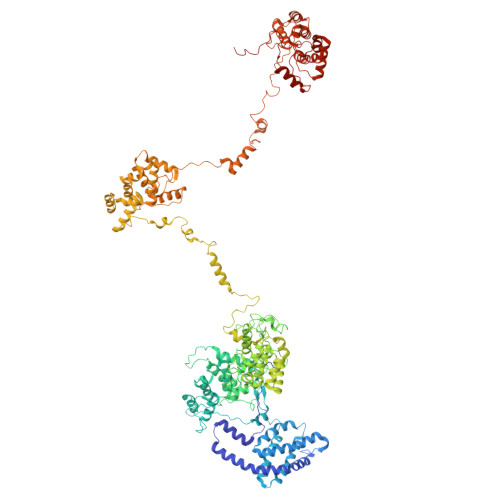
Structure and quenching of a bundle-shaped phycobilisome.
Burtseva, A.D., Slonimskiy, Y.B., Baymukhametov, T.N., Sinetova, M.A., Gvozdev, D.A., Tsoraev, G.V., Cherepanov, D.A., Maksimov, E.G., Popov, V.O., Boyko, K.M., Sluchanko, N.N.(2025) Sci Adv 11: eadz6774-eadz6774
- PubMed: 41091862
- DOI: https://doi.org/10.1126/sciadv.adz6774
- Primary Citation of Related Structures:
9V7G, 9V7H, 9V7I, 9V7J, 9V7K, 9V7L - PubMed Abstract:
Cyanobacteria use soluble antenna megacomplexes, phycobilisomes (PBSs), to maximize light-harvesting efficiency and small photoswitchable orange carotenoid proteins (OCPs) to down-regulate PBSs in high light. Among known PBS morphologies, the one from the basal cyanobacterial genus Gloeobacter still lacks detailed structural characterization. Here, we reconstructed a cryo-electron microscopy structure of the >10-megadalton Gloeobacter violaceus PBS, with diverging, conformationally mobile bundles of rods composed of stacked phycoerythrin and phycocyanin hexamers, stemming from a pentacylindrical allophycocyanin core belted by auxiliary phycocyanin hexamers. We show how two Gloeobacter -specific multidomain linker proteins, Glr1262 and Glr2806, maintain this bundle-shaped architecture and reveal its differential regulation via nonphotochemical quenching by two OCP types of G. violaceus that recognize separate binding sites within the allophycocyanin core, including lateral cylinders absent in tricylindrical cores.
- A.N. Bach Institute of Biochemistry, Federal Research Center of Biotechnology of the Russian Academy of Sciences, Moscow 119071, Russia.
Organizational Affiliation: