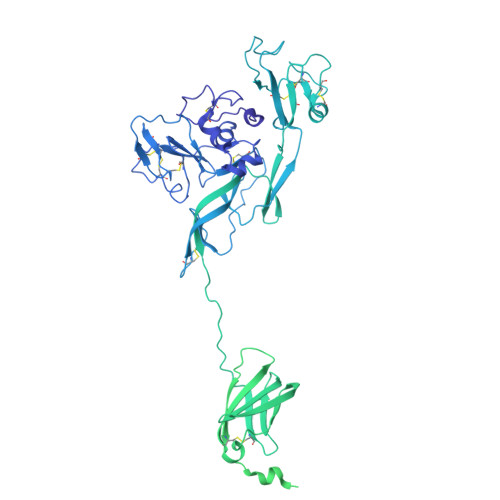
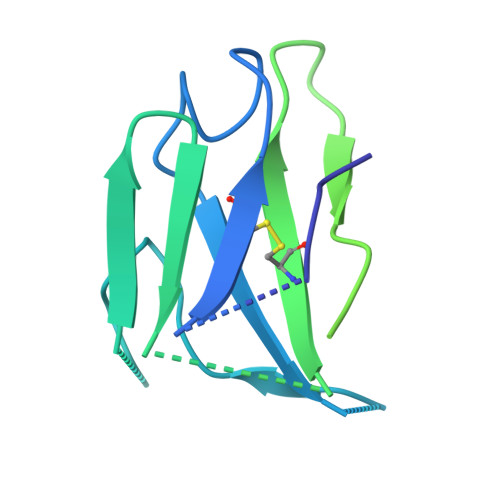
Cryo-EM structure of the Rift Valley fever virus envelope protein in complex with a potent neutralization antibody.
Zhang, L., Meng, K., Xiang, Y.(2025) Proc Natl Acad Sci U S A 122: e2514862122-e2514862122
- PubMed: 41401007
- DOI: https://doi.org/10.1073/pnas.2514862122
- Primary Citation of Related Structures:
9V6N, 9V6R - PubMed Abstract:
Entry of Rift Valley fever virus (RVFV) into host cells is mediated by the viral glycoproteins Gn and Gc. Structural details and assembly mechanism of Gn and Gc on the surface of RVFV remain unclear. Here, we stabilized the GnGc with the neutralizing monoclonal antibody RVFV-140 and determined a near-atomic resolution structure of the GnGc hexamer in complex with the fragment antigen binding (Fab) domain of RVFV-140 (Fab140). Our structure showed that RVFV-140 recognizes a ternary epitope and crosslinks two adjacent Gn heads within the hexamer, thus preventing the prefusion to postfusion transition of the glycoproteins. The intraglycoprotein and interglycoprotein interactions within the GnGc hexamer involve van der Waals forces and hydrogen bonds, which are mainly located between Gn heads, and Gn domain C and Gc domain III. Assembly of the GnGc hexamer requires dramatic conformational changes in the loops L231-L244, D280-Q286, Q380-D386, and A427-Y429 of Gn and the hinge region between domains I and II of Gc. The construction of viral capsomeres with the hexameric structure of recombinant GnGc shows that the capsomeres interact primarily through residues 693 to 713 in domain I of Gc. These interactions vary depending on the local environment of each capsomere.
- Beijing Frontier Research Center for Biological Structure, Center for Infection Biology, School of Basic Medical Sciences, Tsinghua University, Beijing 100084, People's Republic of China.
Organizational Affiliation: