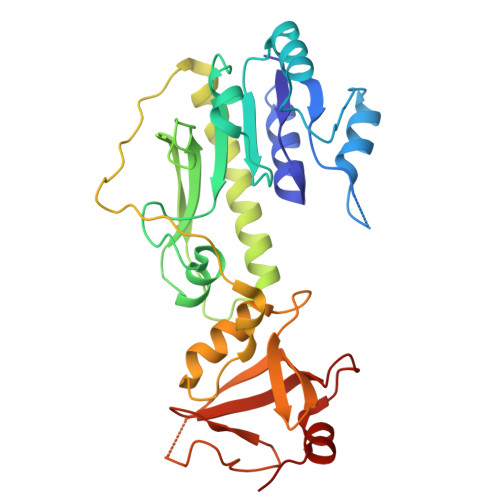
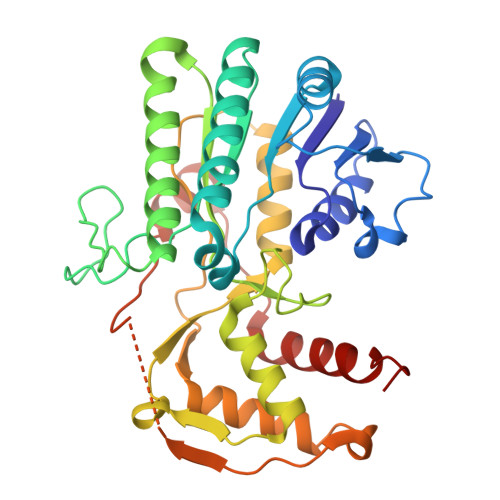
Off-target structural insights: ArnA and AcrB in bacterial membrane-protein cryo-EM analysis.
Caliseki, M., Borucu, U., Yadav, S.K.N., Schaffitzel, C., Kabasakal, B.V.(2025) Acta Crystallogr D Struct Biol 81: 545-557
- PubMed: 40927951
- DOI: https://doi.org/10.1107/S2059798325007089
- Primary Citation of Related Structures:
9V5H, 9V5R - PubMed Abstract:
Membrane-protein quality control in Escherichia coli involves coordinated actions of the AAA+ protease FtsH, the insertase YidC and the regulatory complex HflKC. These systems maintain proteostasis by facilitating membrane-protein insertion, folding and degradation. To gain structural insights into a putative complex formed by FtsH and YidC, we performed single-particle cryogenic electron microscopy on detergent-solubilized membrane samples, from which FtsH and YidC were purified using Ni-NTA affinity and size-exclusion chromatography. Although SDS-PAGE analysis indicated high purity of these proteins, cryo-EM data sets unexpectedly yielded high-resolution structures of ArnA and AcrB at 4.0 and 2.9 Å resolution, respectively. ArnA is a bifunctional enzyme involved in lipid A modification and polymyxin resistance, while AcrB is a multidrug efflux transporter of the AcrAB-TolC system. ArnA and AcrB, known Ni-NTA purification contaminants, were also consistently detected by mass spectrometry in Strep-Tactin affinity-purified samples, validating their presence independently of affinity-tag selection. ArnA, which is typically cytoplasmic, was consistently found in membrane-isolated samples, indicating an association with membrane components. Only 2D class averages corresponding to the cytoplasmic AAA+ domain of FtsH were observed; neither side views of full-length FtsH nor densities corresponding to an intact FtsH-YidC complex could be identified, due to the conformational flexibility of the FtsH complex and its transient interaction with YidC, which limited particle alignment and stable classification in cryo-EM data sets. Two-dimensional class averages revealed additional particles resembling GroEL and cytochrome bo 3 oxidase. These results underscore the utility of cryo-EM in uncovering off-target yet structurally well defined complexes, which may reflect physiologically relevant interactions or purification biases during membrane-protein overexpression.
- Turkish Accelerator and Radiation Laboratory, 06830 Ankara, Türkiye.
Organizational Affiliation: