Structure and function of the nairovirus cap-snatching endonuclease
Kuang, W., Tian, Z., Zhang, G., Wu, F., Li, J., Tang, J., Zhang, H., Zhuo, X., Hu, Z., Wang, M., Zhao, H., Deng, Z.(2025) Nucleic Acids Res
Experimental Data Snapshot
Starting Model: in silico
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RNA-directed RNA polymerase L | A, D [auth B] | 319 | Orthonairovirus haemorrhagiae | Mutation(s): 0 EC: 2.7.7.48 (PDB Primary Data), 3.4.19.12 (PDB Primary Data) | 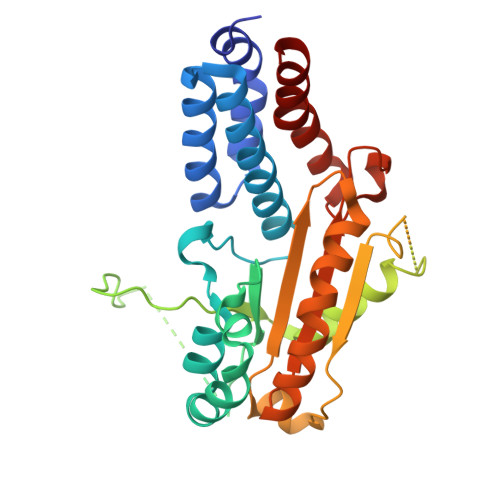 |
UniProt | |||||
Find proteins for D5MEH7 (Orthonairovirus haemorrhagiae) Explore D5MEH7 Go to UniProtKB: D5MEH7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | D5MEH7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| mAb G5 Fab heavy chain | B [auth H], E [auth C] | 228 | Mus musculus | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| mAb G5 Fab light chain | C [auth L], F [auth D] | 219 | Mus musculus | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 98.767 | α = 90 |
| b = 99.779 | β = 90 |
| c = 180.419 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| XDS | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |