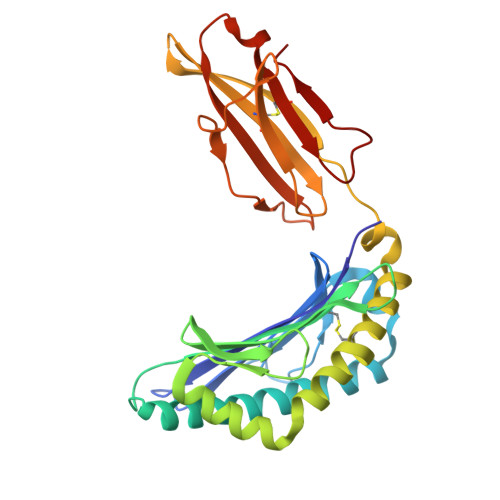
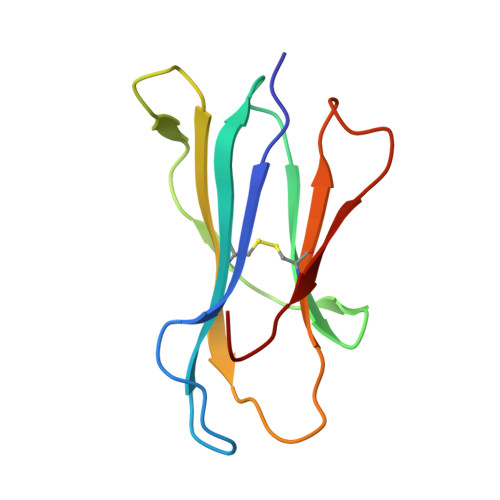
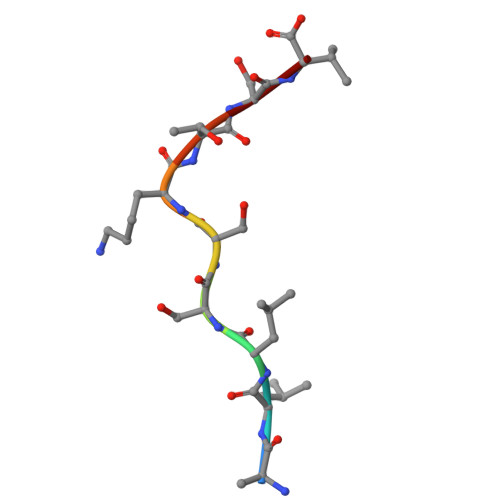
Structural basis of Tibetan wild boar SLA-1*Z0301 reveals conserved peptide presentation and potential high-altitude adaptation.
Fan, S., Kang, C., Peng, J., Wang, T., Ren, S., Li, J., Li, L., Wu, C., Wang, Y., Li, L.(2025) Int J Biol Macromol 320: 145933-145933
- PubMed: 40653219
- DOI: https://doi.org/10.1016/j.ijbiomac.2025.145933
- Primary Citation of Related Structures:
9UL1 - PubMed Abstract:
Major Histocompatibility Complex (MHC) class I molecules facilitate antiviral immunity through their polymorphic peptide-binding diversity. Tibetan wild boars, adapted to extreme plateau conditions, exhibit enhanced disease resistance. Here, we present the first three-dimensional structure of the Tibetan wild boar SLA-1*Z0301 in complex with the GP3-ALL9 peptide derived from porcine reproductive and respiratory syndrome virus (PRRSV). Using X-ray crystallography and AlphaFold3 modeling, we characterized the peptide-binding motif of SLA-1*Z0301, which shares conserved binding pockets with SLA-1*08 alleles found in various domestic pigs, indicating convergent peptide presentation across wild and domestic swine lineages. Thermal stability assays revealed that the primary anchors of SLA-1*Z0301-P2 (B pocket, Tm = 42.5 °C) and P9 (F pocket, Tm = 47.6 °C)-exhibited greater stability compared to mutants in the Heishan pig SLA-3*hs0202 (P3-A = 40.0 °C, P9-A = 38.3 °C), suggesting structural adaptations to high-altitude conditions. A genome-wide screening identified 125 nonapeptides from PRRSV-1 and PRRSV-2 that conform to the SLA-1*Z0301 motif, spanning structural (GP2-GP5) and nonstructural proteins. Notably, five epitopes overlapped with immunodominant responses observed in Large White and Yorkshire × Landrace, highlighting conserved antigen presentation across wild and domestic swine lineages. These results provide valuable insights into the development of cross-species antiviral strategies and vaccine design.
- College of Life Sciences and Agronomy, Zhoukou Normal University, Zhoukou, China; Institute of Neuroscience and Translational Medicine, Zhoukou Normal University, Zhoukou, China. Electronic address: fanshuhuayan@cau.edu.cn.
Organizational Affiliation: