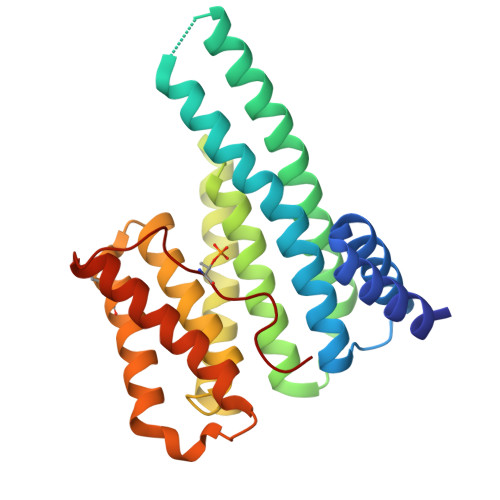
High-resolution structure reveals enhanced 14-3-3 binding by a mutant SARS-CoV-2 nucleoprotein variant with improved replicative fitness.
Perfilova, K.V., Matyuta, I.O., Minyaev, M.E., Boyko, K.M., Cooley, R.B., Sluchanko, N.N.(2025) Biochem Biophys Res Commun 767: 151915-151915
- PubMed: 40318379
- DOI: https://doi.org/10.1016/j.bbrc.2025.151915
- Primary Citation of Related Structures:
9UJ2 - PubMed Abstract:
Replication of many viruses depends on phosphorylation of viral proteins by host protein kinases and subsequent recruitment of host protein partners. The nucleoprotein (N) of SARS-CoV-2 is heavily phosphorylated and recruits human phosphopeptide-binding 14-3-3 proteins early in infection, which is reversed prior to nucleocapsid assembly in new virions. Among the multiple phosphosites of N, which are particularly dense in the serine/arginine-rich interdomain region, phospho-Thr205 is highly relevant for 14-3-3 recruitment by SARS-CoV-2 N. The context of this site is mutated in most SARS-CoV-2 variants of concern. Among mutations that increase infectious virus titers, the S202R mutation (B.1.526 Iota) causes a striking replication boost (∼166-fold), although its molecular consequences have remained unclear. Here, we show that the S202R-mutated N phosphopeptide exhibits a 5-fold higher affinity for human 14-3-3ζ than the Wuhan variant and we rationalize this effect by solving a high-resolution crystal structure of the complex. The structure revealed an enhanced 14-3-3/N interface contributed by the Arg202 side chain that, in contrast to Ser202, formed multiple stabilizing contacts with 14-3-3, including water-mediated H-bonds and guanidinium pi-pi stacking. These findings provide a compelling link between the replicative fitness of SARS-CoV-2 and the N protein's affinity for host 14-3-3 proteins.
- A.N. Bach Institute of Biochemistry, Federal Research Center of Biotechnology of the Russian Academy of Sciences, Moscow, 119071, Russia.
Organizational Affiliation: