Response to Matter Arising: Substrate selection and transition state support the mechanism for GTP-mediate RNA capping catalyzed by SARS-CoV-2 NiRAN
Huang, Y., Tan, L., Liu, Y., Yan, L., Rao, Z., Lou, Z.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RNA-directed RNA polymerase nsp12 | 932 | Severe acute respiratory syndrome coronavirus 2 | Mutation(s): 0 Gene Names: rep, 1a-1b EC: 2.7.7.48 (PDB Primary Data), 2.7.7.50 (PDB Primary Data) | 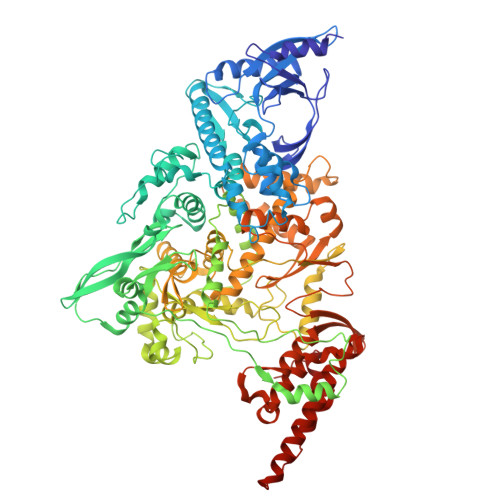 | |
UniProt | |||||
Find proteins for P0DTD1 (Severe acute respiratory syndrome coronavirus 2) Explore P0DTD1 Go to UniProtKB: P0DTD1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0DTD1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Non-structural protein 8 | 198 | Severe acute respiratory syndrome coronavirus 2 | Mutation(s): 0 Gene Names: rep, 1a-1b | 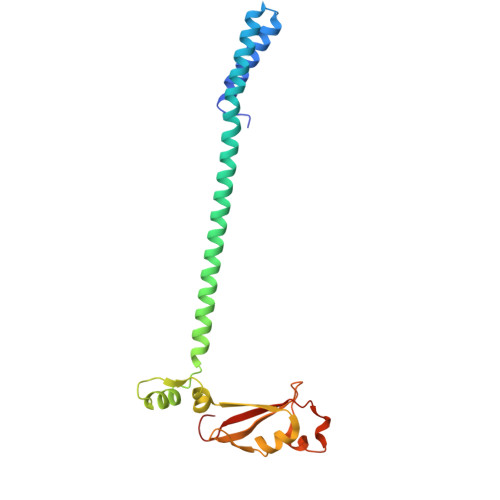 | |
UniProt | |||||
Find proteins for P0DTD1 (Severe acute respiratory syndrome coronavirus 2) Explore P0DTD1 Go to UniProtKB: P0DTD1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0DTD1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Non-structural protein 7 | 78 | Severe acute respiratory syndrome coronavirus 2 | Mutation(s): 0 Gene Names: rep, 1a-1b | 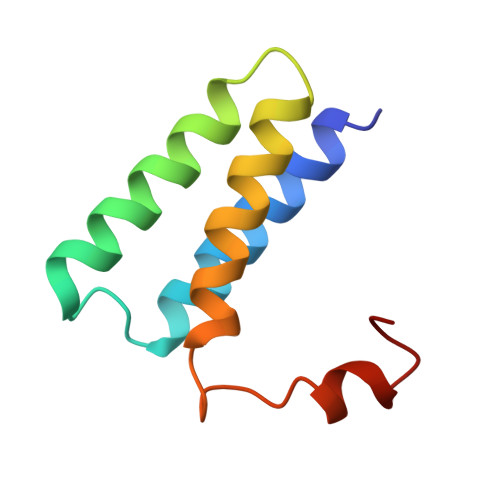 | |
UniProt | |||||
Find proteins for P0DTD1 (Severe acute respiratory syndrome coronavirus 2) Explore P0DTD1 Go to UniProtKB: P0DTD1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0DTD1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Helicase nsp13 | 593 | Severe acute respiratory syndrome coronavirus 2 | Mutation(s): 0 Gene Names: rep, 1a-1b EC: 3.6.4.12 (PDB Primary Data), 3.6.4.13 (PDB Primary Data) | 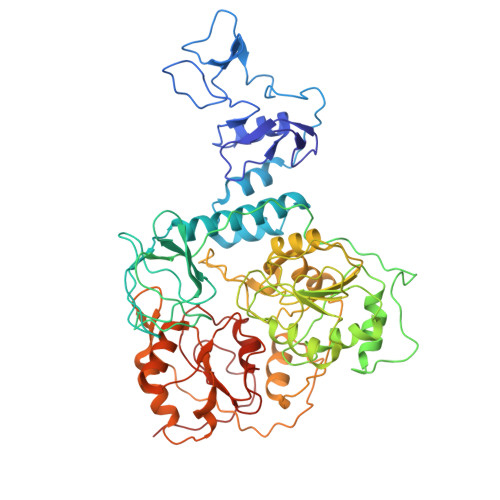 | |
UniProt | |||||
Find proteins for P0DTD1 (Severe acute respiratory syndrome coronavirus 2) Explore P0DTD1 Go to UniProtKB: P0DTD1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0DTD1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Viral protein genome-linked nsp9 | 113 | Severe acute respiratory syndrome coronavirus 2 | Mutation(s): 0 Gene Names: rep, 1a-1b |  | |
UniProt | |||||
Find proteins for P0DTD1 (Severe acute respiratory syndrome coronavirus 2) Explore P0DTD1 Go to UniProtKB: P0DTD1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0DTD1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| RNA (5'-R(P*AP*UP*UP*A)-3') | 4 | Severe acute respiratory syndrome coronavirus 2 |  | ||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| PRIMER | 25 | Severe acute respiratory syndrome coronavirus 2 |  | ||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| TEMPLATE | 27 | Severe acute respiratory syndrome coronavirus 2 |  | ||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GNP (Subject of Investigation/LOI) Query on GNP | M [auth A] | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER C10 H17 N6 O13 P3 UQABYHGXWYXDTK-UUOKFMHZSA-N |  | ||
| ZN Query on ZN | K [auth A] L [auth A] N [auth E] O [auth E] P [auth E] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | -- |