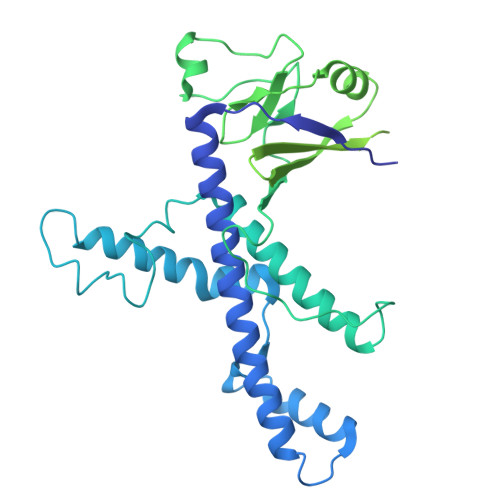
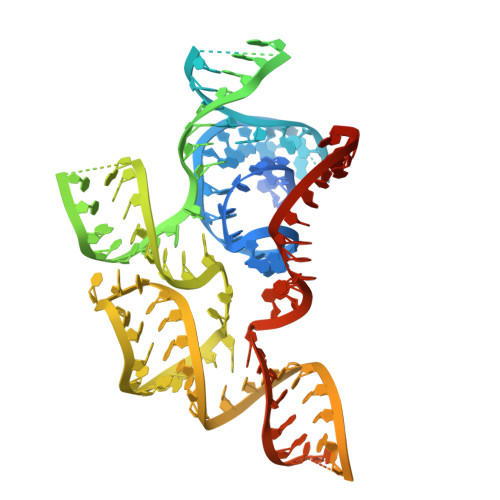
Mechanisms and engineering of a miniature type V-N CRISPR-Cas12 effector enzyme.
Fu, W., Ma, J., Wang, Z., Tang, N., Pan, D., Su, M., Wu, Z., Gan, J., Ji, Q.(2025) Nat Commun 16: 5667-5667
- PubMed: 40595633
- DOI: https://doi.org/10.1038/s41467-025-61290-3
- Primary Citation of Related Structures:
9J09, 9UDI - PubMed Abstract:
Type V CRISPR-Cas12 systems are highly diverse in their functionality and molecular compositions, including miniature Cas12f1 and Cas12n genome editors that provide advantages for efficient in vivo therapeutic delivery due to their small size. In contrast to Cas12f1 nucleases that utilize a homodimer structure for DNA targeting and cleavage with a preference for T- or C-rich PAMs, Cas12n nucleases are likely monomeric proteins and uniquely recognize rare A-rich PAMs. However, the molecular mechanisms behind RNA-guided genome targeting and cleavage by Cas12n remain unclear. Here, we present the cryo-electron microscopy (cryo-EM) structure of Rothia dentocariosa Cas12n (RdCas12n) bound to a single guide RNA (sgRNA) and target DNA, illuminating the intricate molecular architecture of Cas12n and its sgRNA, as well as PAM recognition and nucleic-acid binding mechanisms. Through structural comparisons with other Cas12 nucleases and the ancestral precursor TnpB, we provide insights into the evolutionary significance of Cas12n in the progression from TnpB to various Cas12 nucleases. Additionally, we extensively modify the sgRNA and convert RdCas12n into an effective genome editor in human cells. Our findings enhance the understanding of the evolutionary mechanisms of type V CRISPR-Cas12 systems and offer a molecular foundation for engineering Cas12n genome editors.
- School of Physical Science and Technology & State Key Laboratory of Advanced Medical Materials and Devices, ShanghaiTech University, Shanghai, China.
Organizational Affiliation: