Structural insights into the human HRD1 ubiquitin ligase complex.
Guo, L., Liu, G., He, J., Jia, X., He, Y., Wang, Z., Qian, H.(2025) Nat Commun 16: 6007-6007
- PubMed: 40593878
- DOI: https://doi.org/10.1038/s41467-025-61143-z
- Primary Citation of Related Structures:
9LWU, 9UAV - PubMed Abstract:
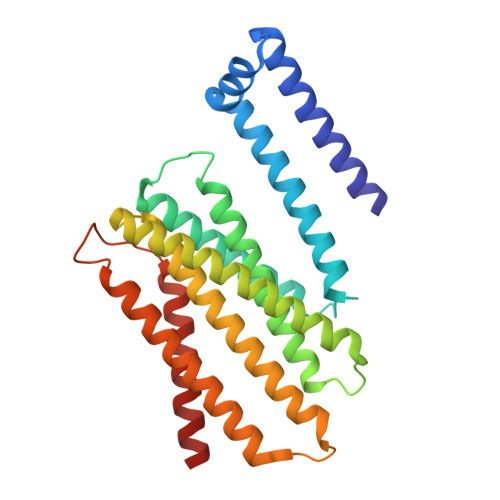
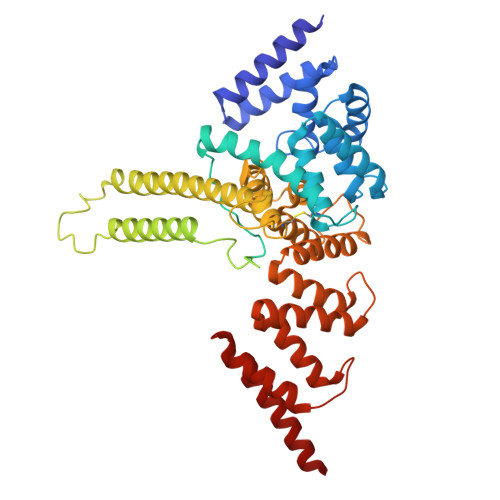
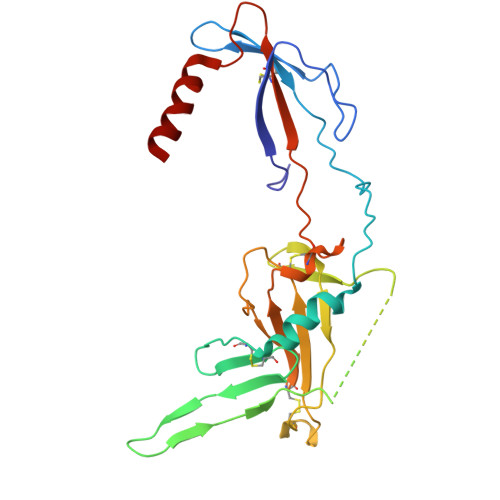
In the endoplasmic reticulum (ER), defective proteins are cleaned via the ER-associated protein degradation (ERAD) pathway. The HRD1 ubiquitin ligase complex, with HRD1, SEL1L, XTP3B or OS9 and Derlin family proteins as the core components, plays essential roles in the recognition, retrotranslocation, and ubiquitination of luminal ERAD substrates. However, the molecular basis is unclear. Here, we determine the cryo-EM structure of the human HRD1-SEL1L-XTP3B complex at 3.3 Å resolution. HRD1 is a dimer, but only one protomer carries the SEL1L-XTP3B complex, forming a 2:1:1 complex. Careful inspection of the EM map reveals a trimmed N-glycan sandwiched by XTP3B and SEL1L, and SEL1L may also contribute to the recognition of the trimmed glycan. The complex undergoes dramatic conformational changes when coexpressed with Derlin proteins. The HRD1 dimer is broken, and two HRD1-SEL1L-XTP3B (1:1:1) units are joined together by a four-helix bundle formed by two SEL1L molecules. The four-helix bundle also touches the micelle, resulting in a bent transmembrane region. These findings indicate that Derlins engagement may induce local curvature in the ER membrane. Cell-based functional assays are conducted to verify the structural observations. Our work provides a structural basis for further mechanistic elucidation of mammalian HRD1 complex-mediated ERAD.
- Department of General Medicine, The First Affiliated Hospital of USTC, MOE Key Laboratory for Membraneless Organelles and Cellular Dynamics, Hefei National Research Center for Interdisciplinary Sciences at the Microscale, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, China.
Organizational Affiliation: