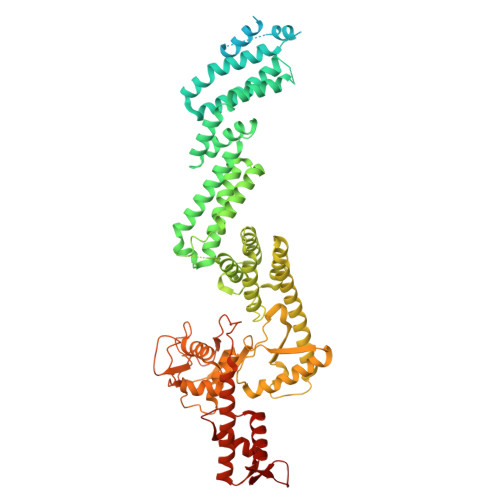
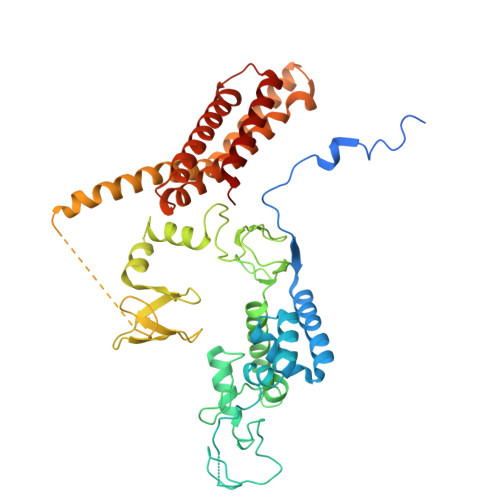
E2 variants for probing E3 ubiquitin ligase activities.
Du, J., Andree, G.A., Horn-Ghetko, D., Stier, L., Singh, J., Kostrhon, S., Kiss, L., Mann, M., Sidhu, S.S., Schulman, B.A.(2026) Proc Natl Acad Sci U S A 123: e2524899122-e2524899122
- PubMed: 41481455
- DOI: https://doi.org/10.1073/pnas.2524899122
- Primary Citation of Related Structures:
9SDX, 9SDY - PubMed Abstract:
E3 ligases partner with E2 enzymes to regulate vast eukaryotic biology. The hierarchical nature of these pairings, with >600 E3s and ~40 E2s in humans, necessitates that E2s cofunction with numerous different E3s. Here, focusing on E3s in the RING-between-RING (RBR) family and their partner UBE2L3 and UBE2D-family E2s, we report an approach to interrogate selected pathways. We screened phage-displayed libraries of structure-based E2 variants (E2Vs) to discover enzymes with enhanced affinity and specificity toward half of all RBR E3 ligases (ARIH1, ARIH2, ANKIB1, CUL9, HOIL1, HOIP, and RNF14). Collectively, these E2Vs allowed distinguishing actions of different cofunctioning E3s, obtaining high-resolution cryogenic Electron Microscopy (cryo-EM) structures of an RBR E3 in the context of a substrate-bound multiprotein complex, and profiling an endogenous RBR E3 response to an extracellular stimulus. Overall, we anticipate that E2V technology will be a generalizable tool to enable in-depth mechanistic and structural analysis of E3 ligase functions, and mapping their activity states and protein partners in cellular signaling cascades.
- Department of Molecular Machines and Signaling, Max Planck Institute of Biochemistry, Martinsried 82152, Germany.
Organizational Affiliation: