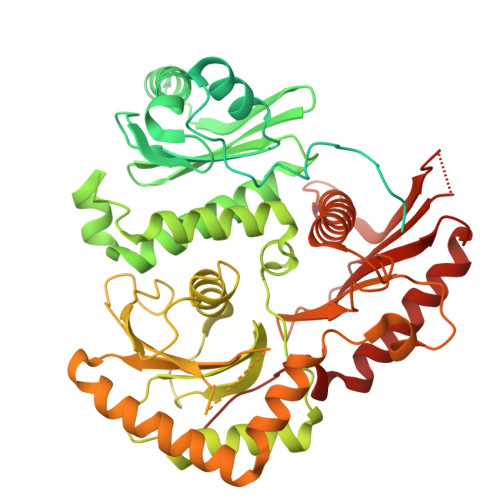
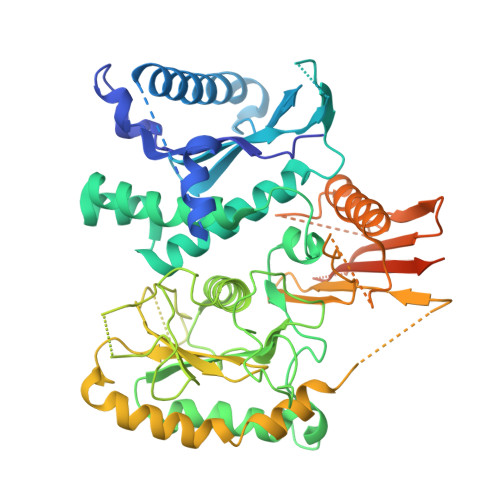
Mechanistic adaptation of the metazoan RabGEFs Mon1-Ccz1 and Fuzzy-Inturned.
Wilmes, S., Tonjes, J., Drechsler, M., Ruf, A., Schafer, J.H., Lurick, A., Januliene, D., Apelt, S., Di Iorio, D., Wegner, S.V., Loose, M., Moeller, A., Paululat, A., Kummel, D.(2025) Sci Adv 11: eadx2893-eadx2893
- PubMed: 40864718
- DOI: https://doi.org/10.1126/sciadv.adx2893
- Primary Citation of Related Structures:
9RS6, 9RS7, 9RS8, 9RS9 - PubMed Abstract:
Rab GTPases organize intracellular trafficking and provide identity to organelles. Their spatiotemporal activation by guanine nucleotide exchange factors (GEFs) is tightly controlled to ensure fidelity. Our structural and functional comparison of the tri-longin domain RabGEFs Mon1-Ccz1 and Fuzzy-Inturned reveals the molecular basis for their target specificity. Both complexes rely on a conserved sequence motif of their substrate GTPases for the catalytic mechanism, while secondary interactions allow discrimination between targets. We also find that dimeric Mon1-Ccz1 from fungi and the metazoan homologs with the additional third subunit RMC1/Bulli bind membranes through electrostatic interactions via distinct interfaces. Protein-lipid interaction studies and functional characterization in flies reveal an essential function of RMC1/Bulli as mediator of GEF complex membrane recruitment. In the case of Fuzzy-Inturned, reconstitution experiments demonstrate that the BAR (Bin-Amphiphysin-Rvs) domain protein CiBAR1 can support membrane recruitment of the GEF. Collectively, our study demonstrates the molecular basis for the adaptation of TLD-RabGEFs to different cellular functions.
- Department of Chemistry and Pharmacy, Institute of Biochemistry, University of Münster, Münster, Germany.
Organizational Affiliation: