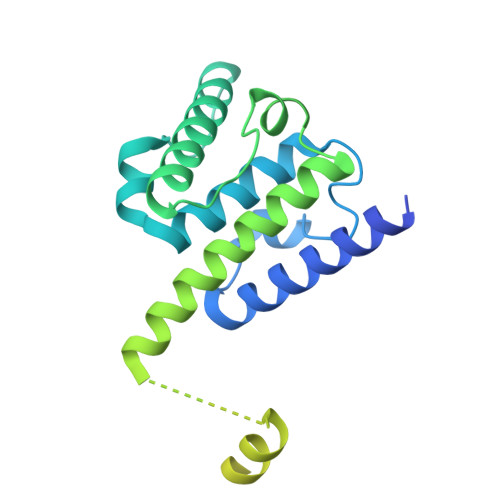
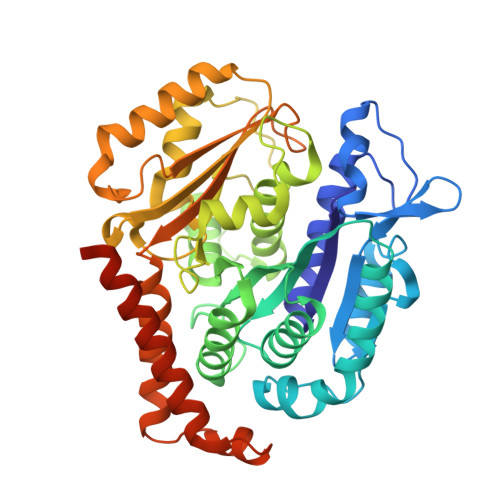
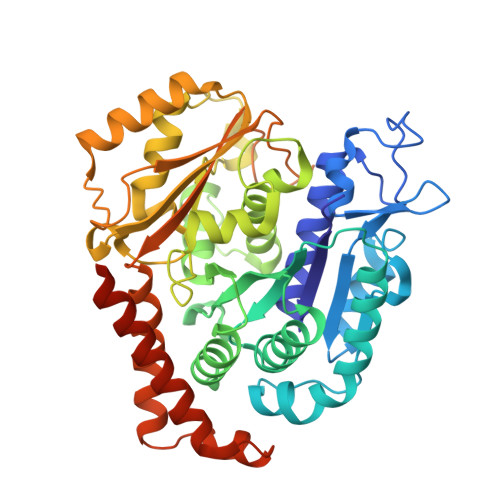
Conserved function of the HAUS6 calponin homology domain in anchoring augmin for microtubule branching.
Wurtz, M., Tonon, G., Vermeulen, B.J.A., Zezlina, M., Gao, Q., Neuner, A., Seidl, A., Konig, M., Harkenthal, M., Eustermann, S., Erhardt, S., Lolicato, F., Schiebel, E., Pfeffer, S.(2025) Nat Commun 16: 7845-7845
- PubMed: 40846850
- DOI: https://doi.org/10.1038/s41467-025-63165-z
- Primary Citation of Related Structures:
9RPD - PubMed Abstract:
Branching microtubule nucleation is a key mechanism for mitotic and meiotic spindle assembly and requires the hetero-octameric augmin complex. Augmin recruits the major microtubule nucleator, the γ-tubulin ring complex, to pre-existing microtubules to direct the formation of new microtubules in a defined orientation. Although recent structural work has provided key insights into the structural organization of augmin, molecular details of its interaction with microtubules remain elusive. Here, we identify the minimal conserved microtubule-binding unit of augmin across species and demonstrate that stable microtubule anchoring is predominantly mediated via the calponin homology (CH) domain in Dgt6/HAUS6. Comparative sequence and functional analyses in vitro and in vivo reveal a highly conserved functional role of the HAUS6 CH domain in microtubule binding. Using cryo-electron microscopy and molecular dynamics simulations in combination with AlphaFold structure predictions, we show that the D. melanogaster Dgt6/HAUS6 CH domain binds microtubules at the inter-protofilament groove between two adjacent β-tubulin subunits and thereby orients augmin on microtubules. Altogether, our findings reveal how augmin binds microtubules to pre-determine the branching angle during microtubule nucleation and facilitate the rapid assembly of complex microtubule networks.
- Zentrum für Molekulare Biologie der Universität Heidelberg (ZMBH), Heidelberg, Germany. m.wuertz@zmbh.uni-heidelberg.de.
Organizational Affiliation: