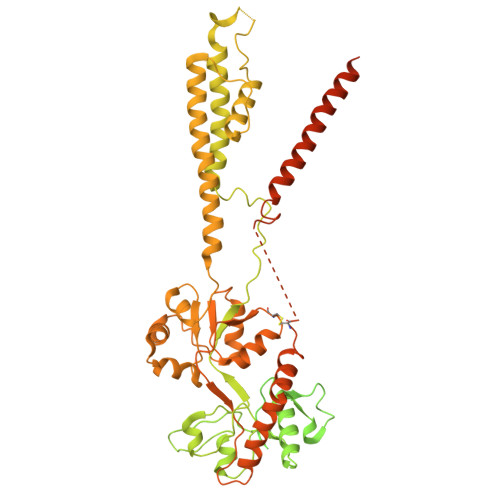
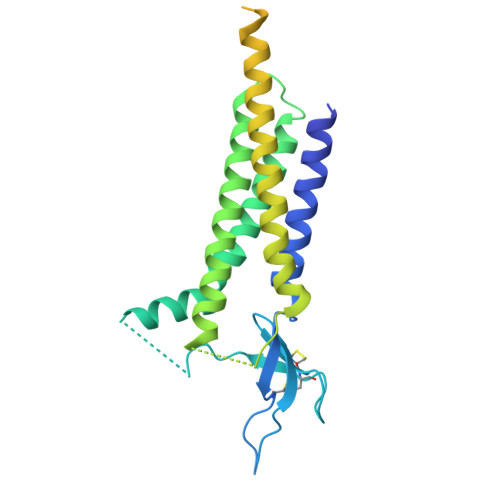
GluA4 AMPA receptor gating mechanisms and modulation by auxiliary proteins.
Vega-Gutierrez, C., Picanol-Parraga, J., Sanchez-Valls, I., Ribon-Fuster, V.D.P., Soto, D., Herguedas, B.(2025) Nat Struct Mol Biol 32: 2416-2428
- PubMed: 40954371
- DOI: https://doi.org/10.1038/s41594-025-01666-7
- Primary Citation of Related Structures:
9IGZ, 9QDN, 9QPW, 9RMS, 9RMW, 9RN4, 9RN7, 9RNH - PubMed Abstract:
AMPA-type glutamate receptors, fundamental ion channels for fast excitatory neurotransmission and synaptic plasticity, contain a GluA tetrameric core surrounded by auxiliary proteins such as transmembrane AMPA receptor regulatory proteins (TARPs) or Cornichons. Their exact composition and stoichiometry govern functional properties, including kinetics, calcium permeability and trafficking. The GluA1-GluA3 subunits predominate in the adult forebrain and are well characterized. However, we lack structural information on full-length GluA4-containing AMPARs, a subtype that has specific roles in brain development and specific cell types in mammals. Here we present the cryo-electron microscopy structures of rat GluA4:TARP-γ2 trapped in active, resting and desensitized states, covering a full gating cycle. Additionally, we describe the structure of GluA4 alone, which displays a classical Y-shaped conformation. In resting conditions, GluA4:TARP-γ2 adopts two conformations, one resembling the desensitized states of other GluA subunits. Moreover, we identify a regulatory site for TARP-γ2 in the ligand-binding domain that modulates gating kinetics. Our findings uncover distinct features of GluA4, highlighting how subunit composition and auxiliary proteins shape receptor structure and dynamics, expanding glutamatergic signaling diversity.
- Institute for Biocomputation and Physics of Complex Systems (BIFI), Department of Biochemistry and Molecular and Cell Biology, University of Zaragoza, Zaragoza, Spain.
Organizational Affiliation: