Structural characterisation of the outer membrane protein SilC, involved in bacterial silver resistance.
Lithgo, R.M., Carr, S.B., Quigley, A.M., Scott, D.J.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Probable outer membrane lipoprotein SilC | 462 | Salmonella enterica subsp. enterica serovar Typhimurium | Mutation(s): 0 Gene Names: silC | 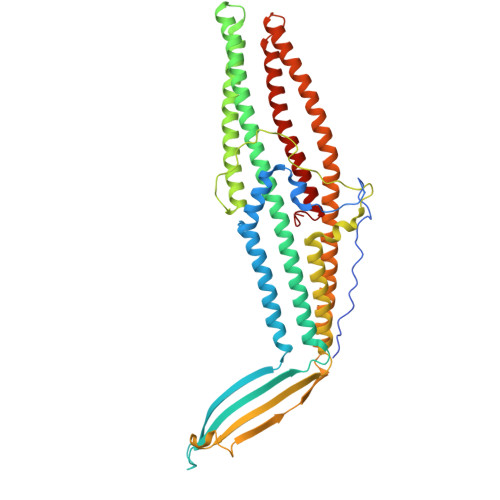 | |
UniProt | |||||
Find proteins for Q9ZHD2 (Salmonella typhimurium) Explore Q9ZHD2 Go to UniProtKB: Q9ZHD2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9ZHD2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Probable outer membrane lipoprotein SilC | 463 | Salmonella enterica subsp. enterica serovar Typhimurium | Mutation(s): 0 Gene Names: silC | 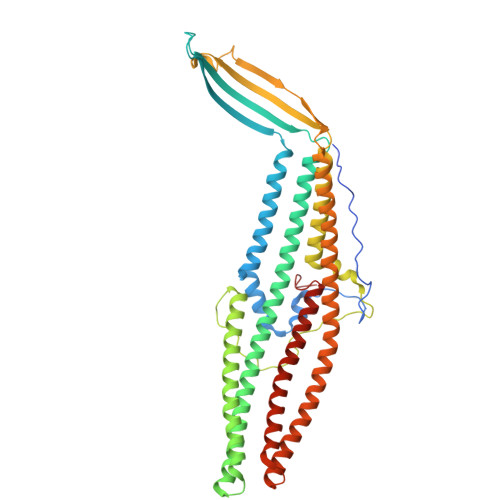 | |
UniProt | |||||
Find proteins for Q9ZHD2 (Salmonella typhimurium) Explore Q9ZHD2 Go to UniProtKB: Q9ZHD2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9ZHD2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 88.055 | α = 90 |
| b = 326.789 | β = 120.27 |
| c = 88.101 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Wellcome Trust | United Kingdom | 202892/Z/16/Z |
| Wellcome Trust | United Kingdom | 223727/Z/21/Z |
| Biotechnology and Biological Sciences Research Council (BBSRC) | United Kingdom | BB/M008770/1 |