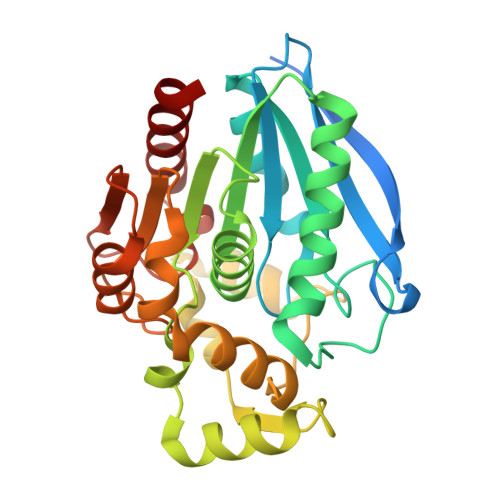
M. tuberculosis meets European lead factory:Identification and structural characterization of novel Rv0183 inhibitors using X-ray crystallography
Riegler-Berket, L., Godl, L., Polidori, N., Aschauer, P., Grininger, C., Prosser, G., Lichtenegger, J., Sagmeister, T., Parigger, L., Gruber, C.C., Reiling, N., Oberer, M.(2025) Disease And Therapeutics 1: 100002