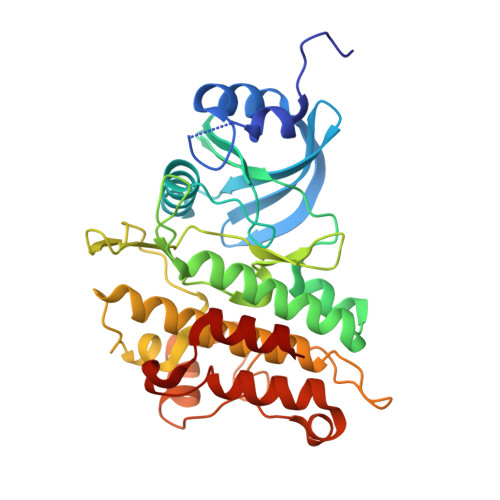
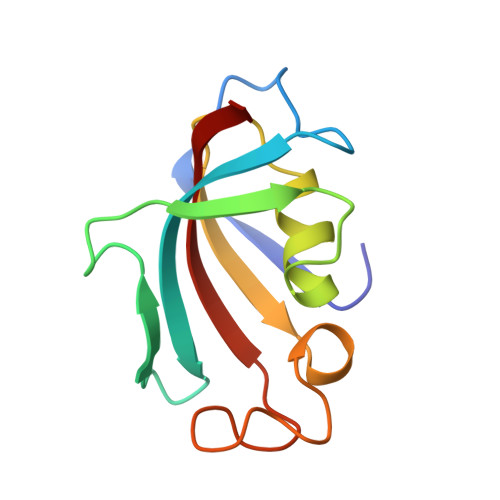
Discovery and Characterization of Zilurgisertib, a Potent and Selective Inhibitor of Activin Receptor-like Kinase‐2 (ALK2) for the Treatment of Fibrodysplasia Ossificans Progressiva.
Ullrich, T., Guth, S., Arista, L., Weiler, S., Stiefl, N., Teixeira-Fouchard, S., Dekker, C., Hinniger, A., Head, V., Kneissel, M., Kramer, I.(2025) ACS Med Chem Lett 16: 2328-2335
- PubMed: 41256990
- DOI: https://doi.org/10.1021/acsmedchemlett.5c00516
- Primary Citation of Related Structures:
9RDA - PubMed Abstract:
Fibrodysplasia ossificans progressiva (FOP) is a rare autosomal-dominant disease leading to progressive soft tissue heterotopic ossification (HO) with no curative treatment available to date. It is caused by gain-of-function mutations in the activin A type-1 receptor ACVR1/ALK2, a member of the bone morphogenetic protein (BMP) type I receptor family. Most recent clinical trials in FOP have adopted for the first time on-target therapies to normalize the aberrant ALK2 receptor activity. Here we describe the discovery and preclinical characterization of zilurgisertib, a novel small-molecule inhibitor of ALK2 kinase with high biochemical and cellular potency, selectivity over other BMP and TGFβ signaling receptor kinases, and excellent oral bioavailability in preclinical species. Zilurgisertib fully suppresses HO in a pediatric mouse model of injury-induced FOP and therefore holds great potential as a novel targeted disease-modifying therapy for FOP. The candidate is being evaluated in clinical trials.
- Global Discovery Chemistry, Novartis Biomedical Research, Novartis Campus, CH-4002 Basel, Switzerland.
Organizational Affiliation: