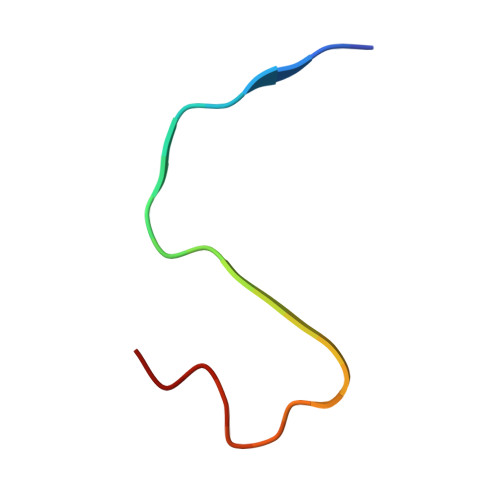
Structural characterization of atrial natriuretic peptide amyloid fibrils from patients with atrial fibrillation.
Broggini, L., Piccoli, M., Chaves-Sanjuan, A., Bonnet, D.M.V., Cirillo, F., Visentin, C., Sonzini, F., Signorelli, P., Lavota, I., Milazzo, M., Nonnis, S., Menicanti, L., Ciconte, G., Pappone, C., Anastasia, L., Ricagno, S.(2025) Nat Commun 16: 9556-9556
- PubMed: 41162370
- DOI: https://doi.org/10.1038/s41467-025-64618-1
- Primary Citation of Related Structures:
9RBD, 9RBW - PubMed Abstract:
Isolated atrial amyloidosis (IAA) is a localized cardiac disorder characterized by atrial natriuretic peptide (ANP) amyloids deposition in the atria, linked to aging and atrial fibrillation (AF). While monomeric ANP regulates blood pressure, its dimeric form is associated with cardiovascular conditions, including AF. The mechanistic link between ANP aggregation, IAA, and AF remains unclear. Here, we present the first high-resolution structural characterization of ANP fibrils extracted from AF patients, revealing two distinct fibril polymorphs. Both present covalent ANP dimers as building blocks but diverge in their structural architecture: one features antiparallel dimers stabilized by a single disulfide bond, while the other consists of parallel dimers bridged by two interchain disulfide bonds. These fibril morphologies were conserved across patients, suggesting a common aggregation mechanism in IAA. Overall, our findings ascribe to dimeric ANP a critical role in amyloid formation, offering promising directions for earlier detection and treatment of IAA.
- Institute for Molecular and Translational Cardiology (IMTC), IRCCS Policlinico San Donato, Piazza Malan, Milan, San Donato Milanese, Italy.
Organizational Affiliation: