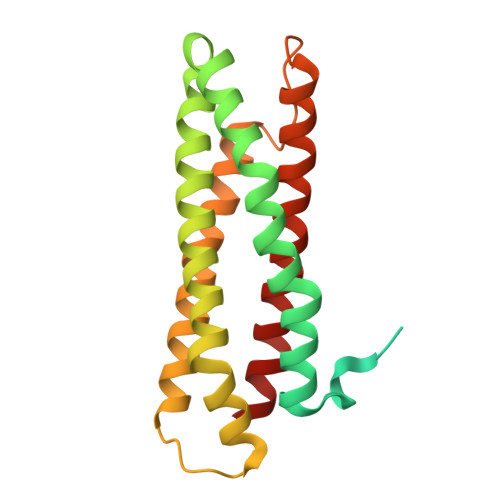
Interaction of Staphylococcus aureus di-iron Repair of Iron Centers protein with the iron-sulfur cluster assembly SUF system.
Lourenco, F.M., Silva, L.S.O., Almeida, M.R., Folgosa, F., Matias, P.M., Carvalho, S.M., Lima, J.C., Romao, C.V., Saraiva, L.M.(2026) Protein Sci 35: e70406-e70406
- PubMed: 41427678
- DOI: https://doi.org/10.1002/pro.70406
- Primary Citation of Related Structures:
9R7J - PubMed Abstract:
Repair of Iron Centers (RIC) proteins are present in several pathogens, including Staphylococcus aureus, contributing to resistance to nitroxidative stress and virulence. In this work, we investigated the molecular mechanisms by which S. aureus RIC facilitates iron mobilization, through structural analyses and functional studies. EPR spectroscopy shows that S. aureus RIC contains an antiferromagnetically coupled di-iron site, undergoing two sequential monoelectronic transitions with a reduction potential of +185 mV. The protein exhibits a stronger affinity for Fe 3+ and releases Fe 2+ more readily, suggesting facilitated Fe 2+ mobilization. The first crystallographic structure for this protein reveals that the di-iron center is embedded in a four-helix bundle domain and is coordinated by histidine residues (H87, H211, H132, H167) and bidentate glutamates (E136, E215). Additionally, we observed that RIC contains one protein tunnel formed by M129 and T170 residues, which are shown to facilitate iron release. In S. aureus, the operon encoding Fe-S biogenesis SUF (SUlFur mobilization) system contains a cysteine desulfurase SufS, a persulfide transporter SufU, and a putative Fe-S scaffold complex SufBC₂D similar to that of Escherichia coli. We show that RIC promotes the assembly of a Fe-S cluster in E. coli SufBC₂D. In S. aureus, we observed the interactions between SufU-SufD and SufC-SufD, which provide the first evidence for the actual formation of this scaffold in the pathogen. Notably, RIC directly interacts with central components of the Fe-S cluster biogenesis, specifically SufS, SufU, and SufD.
- Instituto de Tecnologia Química e Biológica António Xavier, Universidade Nova de Lisboa, Oeiras, Portugal.
Organizational Affiliation: