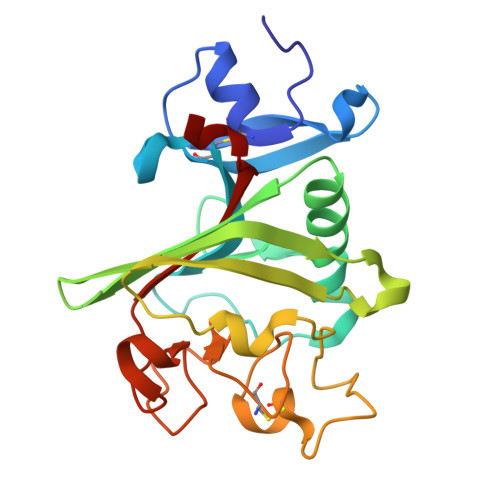
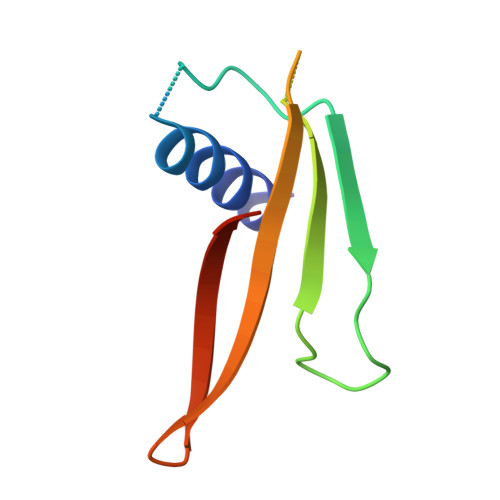
Targeting Tenascin-C-Toll-like Receptor 4 signalling with Adhiron-derived small molecules - a viable strategy for reducing fibrosis in systemic sclerosis.
Gaule, T., Simmons, K.J., Walker, K., Del Galdo, F., Ross, R.L., Viswambharan, H., Krishnappa, J., Pacey, J., McPhillie, M., Tomlinson, D.C., Maqbool, A.(2025) Bioorg Chem 167: 109251-109251
- PubMed: 41289910
- DOI: https://doi.org/10.1016/j.bioorg.2025.109251
- Primary Citation of Related Structures:
9R5Y - PubMed Abstract:
Tissue fibrosis is a hallmark of systemic sclerosis (SSc) and results from the persistent activation of fibroblasts and excessive accumulation of extracellular matrix component such as collagen. Recent evidence implicates the matricellular protein Tenascin-C (TNC) in promoting self-sustaining fibroblast activation and fibrosis via its interaction with Toll-like receptor 4 (TLR4). In this study, we utilized Adhiron-guided ligand discovery to identify small molecule inhibitors targeting the fibrinogen-like globe domain of TNC, a key mediator of TLR4 activation. Two lead compounds (464 and 830) demonstrated structural similarity, favourable ADME profiles, and robust anti-fibrotic activity in vitro. Treatment of dermal fibroblasts derived from SSc patients with either compound significantly reduced Transforming growth factor-β-induced expression of fibrotic genes, ACTA2, COL1A1, COL1A2, and CCN2, and inhibited myofibroblast differentiation. These studies may facilitate the development of effective targeted therapy for fibrosis in SSc and support this novel strategy for small molecule development.
- Clinical Population and Sciences Department, Leeds Institute for Cardiovascular and Metabolic Medicine, School of Medicine, University of Leeds, Leeds, United Kingdom.
Organizational Affiliation: