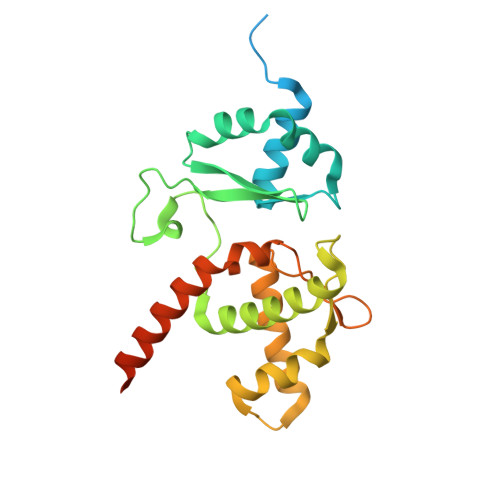
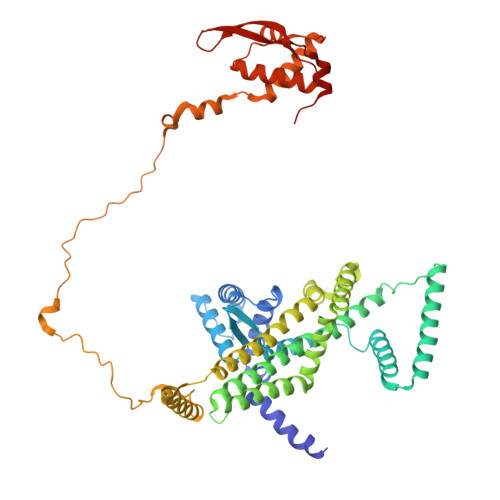
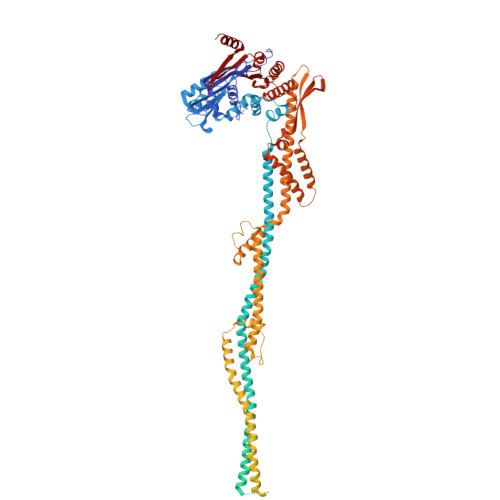
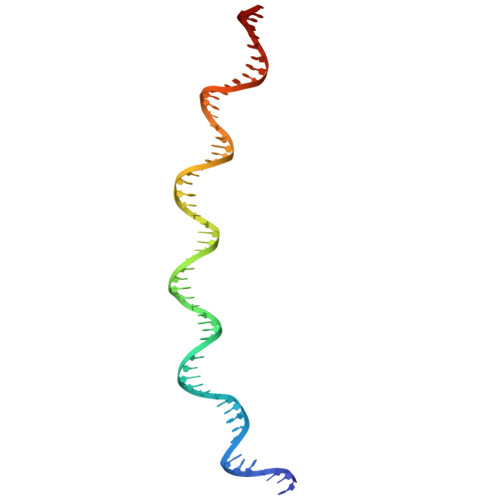
Mechanism of DNA entrapment by a loop-extruding Wadjet SMC motor.
Roisne-Hamelin, F., Liu, H.W., Marechal, N., Uchikawa, E., Durand, A., Gruber, S.(2025) Mol Cell 85: 3898
- PubMed: 41072419
- DOI: https://doi.org/10.1016/j.molcel.2025.09.015
- Primary Citation of Related Structures:
9QXR, 9QXS, 9QXT, 9QXU, 9QXV, 9QXX - PubMed Abstract:
Structural maintenance of chromosome (SMC) complexes perform critical functions by folding DNA through loop extrusion. The choreography and outcome of SMC DNA loading prior to loop extrusion, however, remain elusive. Here, we use cryo-electron microscopy to determine structures of the prokaryotic SMC Wadjet undergoing DNA loading. We show that an initial ATP-triggered relocation of both SMC dimers exposes a DNA-binding pocket and aligns two opened motor units on a DNA double helix. Subsequent ATP hydrolysis drives a nearly 360° rotation of each SMC dimer, closing the motor units around DNA in a sequential manner. This process leads to a DNA-holding conformation-an anticipated key intermediate in loop extrusion-with the DNA held within the kleisin/KITE sub-compartment. Our findings elucidate the mechanism of topological DNA loading by an SMC motor, revealing a straight DNA double helix with motor units oriented tail-to-tail in DNA-holding conformations as the likely starting point of DNA loop extrusion.
- Department of Fundamental Microbiology (DMF), Faculty of Biology and Medicine (FBM), University of Lausanne (UNIL), 1015 Lausanne, Switzerland.
Organizational Affiliation: