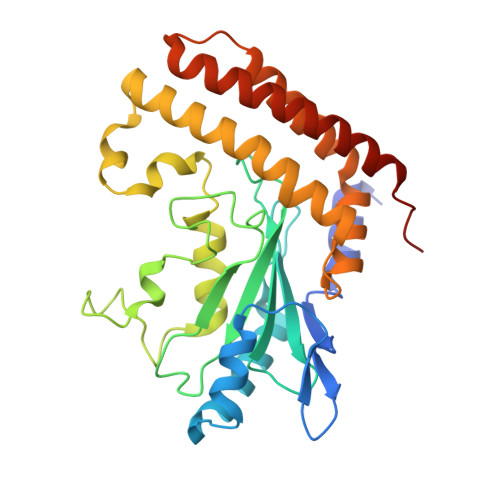
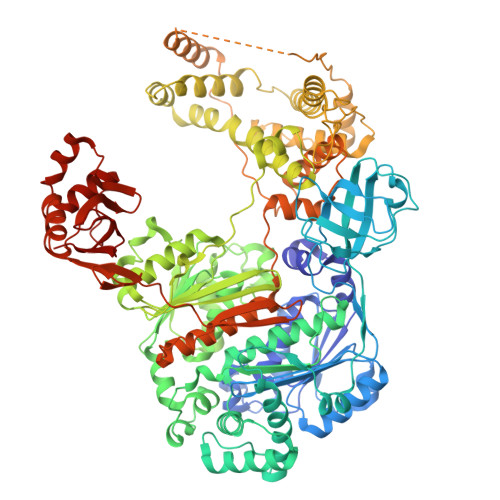
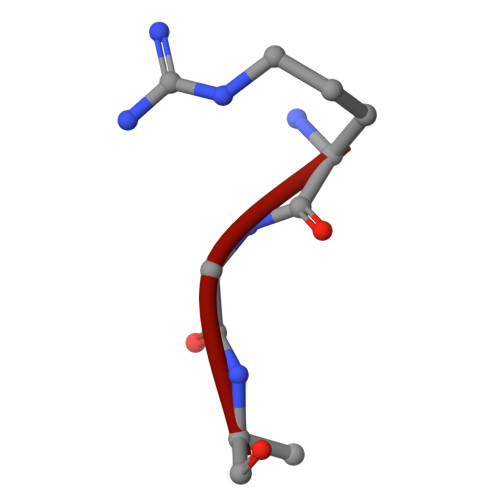
UBA6 specificity for ubiquitin E2 conjugating enzymes reveals a priority mechanism of BIRC6.
Riechmann, C., Ellison, C.J., Anderson, J.W., Hofmann, K., Sarkies, P., Elliott, P.R.(2025) Nat Struct Mol Biol
- PubMed: 41350950
- DOI: https://doi.org/10.1038/s41594-025-01717-z
- Primary Citation of Related Structures:
9QGG, 9QGI, 9QGR, 9QGW, 9QH5, 9QHI, 9QIA, 9QIC, 9QIG, 9QII, 9QIM, 9QIO, 9QIP, 9QIV - PubMed Abstract:
In mammals, ubiquitylation is orchestrated by the canonical ubiquitin-activating E1 enzyme UBA1 and the orthogonal E1 UBA6. Growing evidence underscores the essentiality of both E1s, which differentiate between 29 active ubiquitin-conjugating enzymes (E2s). The mechanisms governing this distinction have remained unclear. Here we establish a framework for ubiquitin E1-E2 specificity. Focusing on UBA6-controlled ubiquitylation cascades, we reveal that BIRC6, a UBA6-exclusive E2, gains priority over all other UBA6-competent E2s, underpinning the functional importance of defined UBA6-BIRC6 ubiquitylation events in regulating cell death, embryogenesis and autophagy. By capturing BIRC6 receiving ubiquitin from UBA6 in different states, we observe BIRC6 engaging with the UBA6 ubiquitin fold domain, driving an exceptionally high-affinity interaction that is modulated by the UBA6 Cys-Cap loop. Using this interaction as a template, we demonstrate how to confer activity between E2s and their noncognate E1, providing a tool to delineate E1-E2-dependent pathways. Lastly, we explain how BIRC6 priority does not lead to inhibition of UBA6, through a bespoke thioester switch mechanism that disengages BIRC6 upon receiving ubiquitin. Our findings propose a concept of hierarchy of E2 activity with cognate E1s, which may explain how ubiquitin E1s can each function with over a dozen E2s and orchestrate E2-specific cellular functions.
- Department of Biochemistry, University of Oxford, Oxford, UK.
Organizational Affiliation: