Structural studies on ribosomes of differentially macrolide-resistant Staphylococcus aureus strains.
Rivalta, A., Fedorenko, A., Le Scornet, A., Thompson, S., Halfon, Y., Breiner Goldstein, E., Cavdaroglu, S., Melenitzky, T., Hiregange, D.G., Zimmerman, E., Bashan, A., Yap, M.F., Yonath, A.(2025) Life Sci Alliance 8
- PubMed: 40490363
- DOI: https://doi.org/10.26508/lsa.202503325
- Primary Citation of Related Structures:
9QEG, 9QEH - PubMed Abstract:
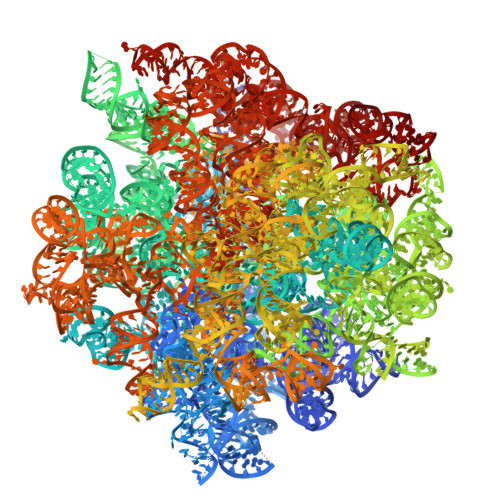
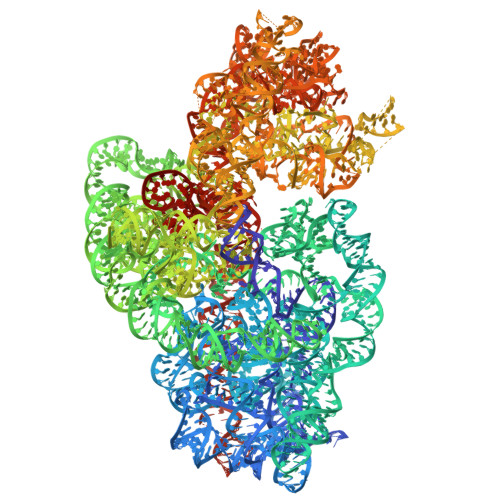
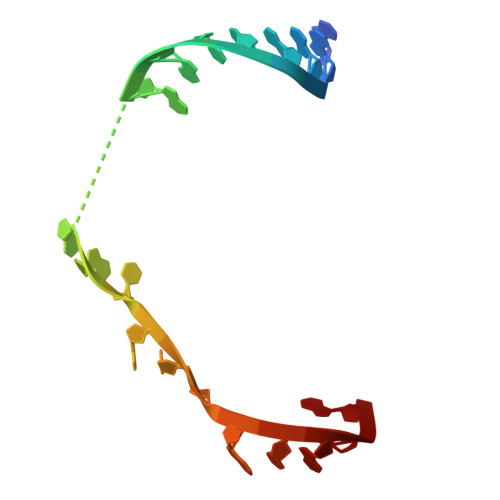
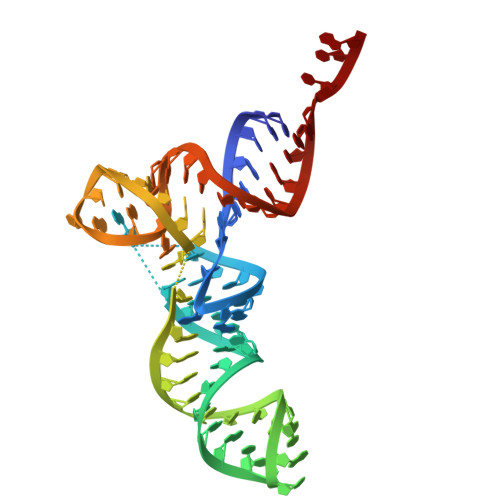
Antimicrobial resistance is a major global health challenge, diminishing the efficacy of many antibiotics, including macrolides. In Staphylococcus aureus , an opportunistic pathogen, macrolide resistance is primarily mediated by Erm-family methyltransferases, which mono- or dimethylate A2058 in the 23S ribosomal RNA, reducing drug binding. Although macrolide-ribosome interactions have been characterized in nonpathogenic species, their structural basis in clinically relevant pathogens remains limited. In this study, we investigate the impact of ermB -mediated resistance on drug binding by analyzing ribosomes from S. aureus strains with varying levels of ermB expression and activity. Using cryo-electron microscopy, we determined the high-resolution structures of solithromycin-bound ribosomes, including those with dimethylated A2058. Our structural analysis reveals the specific interactions that enable solithromycin binding despite double methylation and resistance, as corroborated by microbiological and biochemical data, suggesting that further optimization of ketolide-ribosome interactions could enhance macrolide efficacy against resistant S. aureus strains.
- Department of Chemical and Structural Biology, Weizmann Institute of Science, Rehovot, Israel.
Organizational Affiliation: